Abstract
Planctomycetes is a phylum of environmentally important bacteria, which also receive significant attention due to their fascinating cell biology. Access to axenic Planctomycete cultures is crucial to study cell biological features within this phylum in further detail. In this study, we characterise three novel strains, Mal52T, Pan258 and CA54T, which were isolated close to the coasts of the islands Mallorca (Spain) and Panarea (Italy), and from Monterey Bay, CA, USA. The three isolates show optimal growth at temperatures between 22 and 24 °C and at pH 7.5, divide by polar budding, lack pigmentation and form strong aggregates in liquid culture. Analysis of five phylogenetic markers suggests that the strains constitute two novel species within a novel genus in the family Planctomycetaceae. The strains Mal52T (DSM 101177T = VKM B-3432T) and Pan258 were assigned to the species Symmachiella dynata gen nov., sp. nov., while strain CA54T (DSM 104301T = VKM B-3450T) forms a separate species of the same genus, for which we propose the name Symmachiella macrocystis sp. nov.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Planctomycetes were first discovered in 1924 and mistakenly acknowledged as eukaryotes (Gimesi 1924), but later reclassified as bacteria (Hirsch 1972). Planctomycetes are ubiquitous bacteria dwelling in marine, limnic and soil environments, in which they play an important role in the global carbon and nitrogen cycle (Wiegand et al. 2018). The eponymous phylum Planctomycetes is part of the PVC superphylum, which additionally includes the phyla Verrucomicrobia, Chlamydiae and other sister phyla. The PVC superphylum has medical and biotechnological relevance (Rivas-Marin and Devos 2018; Wagner and Horn 2006). According to the current taxonomy, the phylum Planctomycetes is divided into the classes Phycisphaerae and Planctomycetia. Candidatus Brocadiae might very well form a third class within the phylum, but no axenic cultures have been obtained from this class so far (Kartal et al. 2013). Known members of the class Planctomycetia divide by budding, while binary fission was observed as cell division mode in the class Phycisphaerae. The class Planctomycetia was recently re-organised and is now further subdivided into the orders Isosphaerales, Gemmatales, Planctomycetales and Pirellulales (Dedysh et al. 2020b).
Planctomycetes can be found in various habitats on earth and can even be amongst the most abundant phyla in bacterial communities on biotic surfaces, e.g. on marine macroscopic phototrophs (Bengtsson and Øvreås 2010; Bondoso et al. 2014, 2015, 2017; Lage and Bondoso 2014). Given the oligotrophic nature of seawater, Planctomycetes are suggested to use complex substrates secreted by phototrophs as sources of carbon and energy (Jeske et al. 2013; Lachnit et al. 2013). Indeed, in silico genome analyses point towards higher numbers of carbohydrate-active enzymes encoded by Planctomycetes (Ivanova et al. 2017; Wallner et al. 2005; Wegner et al. 2013). In this context, pili originating from crateriform structures and an enlarged periplasmic space are discussed to be part of a specific uptake system, which may allow intracellular digestion of entire high-molecular weight sugar molecules (Boedeker et al. 2017). If true, this strategy is a decisive advantage over the use of extracellular enzymes for degradation since the latter strategy would provide easily degradable carbon sources to competitors.
Despite the assumed presence of such catabolic systems, the high abundance of Planctomycetes is still unexpected given their slow growth compared to many other heterotrophic bacteria competing with Planctomycetes for ‘nutrient-rich’ ecological niches (Frank et al. 2014; Wiegand et al. 2018). The potential for production of small molecules with antimicrobial properties may also play a decisive role in such environments (Graça et al. 2016; Jeske et al. 2013).
Morphologically, Planctomycetes have been suggested to possess uncommon traits compared to canonical bacteria. Different traits, including the lack of peptidoglycan (König et al. 1984), a compartmentalised cell plan (Lindsay et al. 1997), a nucleus-like structure (Fuerst and Webb 1991) and endocytosis-like uptake (Lonhienne et al. 2010) have been proposed. Some of these traits were found not to be entirely accurate. The compartmentalised cell plan turned out to be invaginations of the cytoplasmic membrane (Acehan et al. 2013; Boedeker et al. 2017), while presence of peptidoglycan was demonstrated (Jeske et al. 2015; Van Teeseling et al. 2015). The cell plan of Planctomycetes was revised based on the use of novel microscopy techniques and genetic tools, and the cell envelope architecture is now considered similar to that of Gram-negative bacteria (Devos 2014; Jogler et al. 2011; Jogler and Jogler 2013; Rivas-Marin et al. 2016). However, Planctomycetes are still unusual. They e.g. lack canonical divisome proteins including the otherwise essential FtsZ (Jogler et al. 2012; Pilhofer et al. 2008) and 40–55% of the proteins encoded in planctomycetal genomes are of unknown function.
For extending the current collection of Planctomycetes available as axenic cultures, here we describe three novel closely related strains, which we isolated from algae close to the island Mallorca, from seagrass leaves close to the island Panarea and from the kelp forest at Monterey Bay in California, USA.
Materials and methods
Isolation of the novel strains
The three novel strains Mal52T, Pan258 and CA54T were isolated as previously described (Wiegand et al. 2020). Strain CA54T was isolated from a Macrocystis pyrifera kelp forest at Monterey Bay, CA, USA on November 28th, 2014 (exact location: 36.619 N 121.901 W). Strain Mal52T was obtained from algae in the Mediterranean Sea close to S’Arenal, Mallorca, Spain (exact location: 39.5126 N 2.7470 E) on September 23rd, 2014. Strain Pan258 was isolated from seagrass leaves growing next to a natural gas escape of the hydrothermal vent system close to Panarea Island (exact location: 38.6457 N 15.0772 E), which were sampled on September 9th, 2013. In order to prevent fungal growth, pieces of kelp, alga and seagrass were initially rinsed with 100 mg/L cycloheximide dissolved in sterile-filtered natural seawater and subsequently swabbed over plates with solidified M1H NAG ASW medium (Kallscheuer et al. 2019a) containing 8 g/L gellan gum, 1000 mg/L streptomycin, 200 mg/L ampicillin and 20 mg/L cycloheximide. The plates were incubated at 20 °C for at least six weeks. Colonies obtained were restreaked on fresh plates, which were used to inoculate liquid M1H NAG ASW medium. Sequencing of the 16S rRNA gene of the colonies was performed according to a previously published protocol to ensure that novel strains are members of the phylum Planctomycetes (Rast et al. 2017).
Light and electron microscopy
Phase contrast and scanning electron microscopic analyses were performed as described in a previous study (Boersma et al. 2019).
Genome information and genome-based analysis of the carbon metabolism
The genome sequences of the three novel isolates are available from GenBank under accession numbers CP036270 (Pan258), CP036276 (Mal52T) and SJPP00000000 (CA54T). The 16S rRNA gene sequences can be found under accession numbers MK554517 (Pan258), MK554513 (Mal52T) and MK554522 (CA54T). DNA isolation and genome sequencing are part of a previous study (Wiegand et al. 2020). The genome-based analysis of the carbon metabolism of the novel isolates was performed as previously described (Rivas-Marin et al. 2020).
Physiological analyses
The pH optimum for growth was determined in M1H NAG ASW medium with 100 mM of the following buffers: 2-(N-morpholino)ethanesulfonic acid (MES) for pH 5.0 and 6.0, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) for pH 7.0, 7.5 and 8.0 and N-cyclohexyl-2-aminoethanesulfonic acid (CHES) for pH 9.0 and 10.0. The cultures were incubated at 28 °C. The temperature optimum for growth was determined by cultivation at temperatures ranging from 10 to 40 °C at pH 8.0. All cultivations were performed in triplicates and growth was assessed by measuring the optical density at 600 nm (OD600). Growth rates for each tested condition were calculated by plotting ln(OD600), the natural logarithm of average OD600 values from biological triplicates, against the cultivation time. The slope of the linear range of this plot (at least five data points) was used as maximal growth rate µmax (in h− 1). Generation times td (in h) were calculated using the formula td = ln(2)/µmax.
Phylogenetic analyses
Maximum likelihood 16S rRNA gene sequence-based phylogeny was computed for the novel strains, the described type strains of all planctomycetal species (as of June 2020), including recently published strains (Boersma et al. 2019; Dedysh et al. 2020a, b; Kallscheuer et al. 2019a, b, c, 2020a, b; Kohn et al. 2020; Peeters et al. 2020; Wiegand et al. 2020). The alignment of 16S rRNA gene sequences was performed with SINA (Pruesse et al. 2012). A maximum likelihood approach with 1000 bootstraps, nucleotide substitution model GTR, gamma distribution and estimation of proportion of invariable sites (Stamatakis 2014) was used. The outgroup consisted of three 16S rRNA gene from strains outside of the phylum Planctomycetes, but still part of the PVC superphylum. For the multi-locus sequence analysis (MLSA), the unique single-copy core genome of the analysed genomes was determined with proteinortho5 (Lechner et al. 2011) with the ‘selfblast’ option enabled. The protein sequences of the resulting orthologous groups were aligned using MUSCLE v.3.8.31 (Edgar 2004). After clipping, partially aligned C- and N-terminal regions and poorly aligned internal regions were filtered using Gblocks (Castresana 2000). The final alignment was concatenated and clustered using the maximum likelihood method implemented by RAxML (Stamatakis 2014) with the ‘rapid bootstrap’ method and 500 bootstrap replicates. Five planctomycetal genomes from the order Pirellulales served as outgroup. The rpoB gene sequences were taken from publicly available online databases and sequence identities were determined as previously described (Bondoso et al. 2013). The average nucleotide identity (ANI) was calculated with OrthoANI (Lee et al. 2016). The average amino acid identity (AAI) was calculated using the aai.rb script of the enveomics collection (Rodriguez-R and Konstantinidis 2016) and percentage of conserved proteins (POCP) was calculated as described (Qin et al. 2014).
Results and discussion
Phylogenetic inference
In the phylogenetic trees obtained after analysis of 16S rRNA genes and MLSA, the strains Mal52T, Pan258 and CA54T form a monophyletic cluster within the family Planctomycetaceae (Fig. 1). Both trees as well as five analysed phylogenetic markers suggest Maioricimonas rarisocia Mal4T (Rivas-Marin et al. 2020) and Gimesia maris (Scheuner et al. 2014) as current closest relatives of the three novel isolates. Based on this finding, we analysed 16S rRNA gene sequence similarity, rpoB gene similarity, AAI and POCP to check whether the novel isolates belong to one of the two genera. The three strains share a minimal 16S rRNA gene sequence identity of 89.1% with M. rarisocia Mal4T and 88.4% with Gimesia sp. Both values are significantly below the proposed genus threshold of 94.5% (Yarza et al. 2014), indicating that these strains belong to a separate, yet undescribed genus in the family Planctomycetaceae (Fig. 2). This finding is also supported by analyses of rpoB similarity, AAI and POCP, since comparison of the three novel isolates with members of the above-mentioned genera yielded values below the respective genus thresholds of 75.5–78% for rpoB (Kallscheuer et al. 2019c), 60% for AAI (Konstantinidis and Tiedje 2005) and approximately 50% for POCP (Qin et al. 2014) (Fig. 2). ANI values in a range of 65–67% and thus far below the species threshold of 95% (Kim et al. 2014) thereby also ensure that the novel strains do not belong to any described species.
Maximum likelihood 16S rRNA gene sequence-(a) and MLSA-based phylogenetic trees (b) depicting the positions of strains Mal52T, Pan258 and CA54T. Phylogeny was calculated as described in the Material and methods section. Bootstrap values after 1000 re-samplings (500 re-sampling for MLSA analysis) are given at the nodes (in %). The tree scale (branch length values) represents the mean expected rates of substitution per site. The outgroups consist of three 16S rRNA genes from the PVC superphylum outside of the phylum Planctomycetes (a) and of members of the order Pirellulales (b)
Next, we compared the strains Mal52T, Pan258 and CA54T against each other to check if they belong to separate species. It turned out that strains Mal52T and Pan258 have a 100% identical 16S rRNA gene sequence, indicating that they belong to the same species. This assumption is supported by an ANI of 96.5% above the species threshold of 95% and an AAI of 97.7% (proposed species threshold of 95–96%) (Konstantinidis and Tiedje 2005). Only the rpoB similarity of 95.6% is below, but still close to the species threshold of 96.3% (Bondoso et al. 2013) (Fig. 2). In particular due to an identical 16S rRNA gene sequence, we conclude that the strains Mal52T and Pan258 belong to the same species. In constrast, comparison of either of these two strains with strain CA54T yielded identity values for AAI and ANI significantly below the species threshold values (Fig. 2). Although strain CA54T shares an identity of 99.5% on 16S rRNA gene sequence level (species threshold 98.7%), we decided to assign it to a separate species. This decision is based on previous observations that this threshold is not always applicable for members of the class Planctomycetia and that strains can belong to separate species despite 16S rRNA gene sequence similarities above the threshold (Kohn et al. 2020). Taken together, the phylogenetic analysis suggests that the three strains represent two novel species of a novel genus within the family Planctomycetaceae.
Morphological and physiological analyses
For microscopic analyses of the three isolated strains, cells were harvested during the exponential growth phase. Detailed information on morphology, cell division and motility is summarised in Table 1. The current closest relatives M. rarisocia and G. maris were chosen for comparison. Strain Mal52T (Figs. 3a–c, 4a,b) and strain Pan258 (Figs. 3d–f, 4c,d) form white colonies on plates and cells have an ovoid to pear-shaped morphology. Strain CA54T displayed white- to cream-coloured colonies. Cells of this strain were ovoid to pear-shaped, but also rod-shaped cells were observed (Figs. 3g–i, 4e,f); a phenotype that was not found for the other two isolates. The cell shape of the novel isolates differs from spherical G. maris cells. While the average cell size of strains Mal52T, Pan258 and CA54T turned out to be similar (1.6–1.8 × 0.8–1.0 µm) (Fig. 3c,f,i), all three are slightly smaller than cells of M. rarisocia Mal4T.
The lack of pigmentation indicates the incapability of the strains to form carotenoids. In that regard, they are similar to G. maris, but differ from the orange pigmentation of M. rarisocia. A strong tendency to aggregate and biofilm formation was observed. This is a considerable difference to M. rarisocia Mal4T, which mostly occurs in the form of single cells and only in rare cases forms aggregates. Crateriform structures could only be observed on the surface of Mal52T cells, however we cannot exclude the presence in case of the other two strains. Cells of all three strains are motile and divide by polar budding.
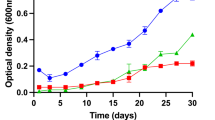
During cultivation experiments, strains Mal52T and Pan258 grew over a temperature range of 15–30 °C and a pH range of 5.5–9.5 (Table 1). Strain CA54T showed a similar pH range and all three strains showed optimal growth at pH 7.5. The optimum temperature for growth falls between 22 and 24 °C and is thus considerably lower than observed for M. rarisocia and G. maris (30-33 °C). The novel isolates are slow-growing strains with maximal growth rates between 0.005 and 0.01 h− 1 (generation times of 70–140 h) in M1H NAG ASW medium.
Genomic characteristics
Genome characteristics are listed in Table 1. The three novel isolates and the two species chosen for comparison have very similar genome sizes of 7.6–7.8 Mb. Not surprisingly, numbers of protein-coding genes (5,829-6,143), protein-coding genes per Mb (753–803) and coding densities (85.4–86.9%) are similar. In contrast, the novel strains can be clearly differentiated from M. rarisocia and G. maris by differences in the DNA G + C content of their genomes (Table 1). Strain Mal52T has two copies of the 16S rRNA gene, while only a single 16S rRNA gene was found in the genomes of the other two novel isolates. None of the compared strains harbors plasmids. In all five genomes 39–41% of the automatically annotated genes code for proteins with unknown function. These values are in the lower range of 40–55% observed in genomes of Planctomycetes sequenced so far.
Genome-based analysis of enzymes participating in the central carbon metabolism
Based on the genomes of strains Pan258, Mal52T and CA54T, the presence of key metabolic enzymes of the central carbon metabolism was analysed. The analysis included glycolytic pathways (Embden–Meyerhof–Parnas pathway or Entner–Doudoroff pathway), the tricarboxylic acid (TCA) cycle, gluconeogenesis and anaplerotic reactions (Table 2). All three strains contain genes coding for enzymes involved in glycolysis, both for the Embden–Meyerhof–Parnas pathway and the Entner–Doudoroff pathway. In addition, key enzymes for sugar degradation via the pentose phosphate pathway were found in all three strains. This was not surprising since important precursors for amino acid and nucleotide biosynthesis branch off from the pentose phosphate pathway and auxotrophies occur in case that this pathway is non-functional. Further analysis showed that genes coding for all enzymes of the TCA cycle could be found in each strain. Genes coding for enzymes required for conversion of oxaloacetate to phosphoenolpyruvate and for C1-dephosphorylation of fructose-1,6-bisphosphate as key steps of a functional gluconeogenesis were identified. Thus, all three strains should be capable of de novo sugar biosynthesis. In contrast, the glyoxylate shunt, an important anaplerotic pathway during growth on acetate or fatty acids, is absent in all three strains, which appears to be a common feature of Planctomycetes.
Taken together, phylogenetic inference as well as morphological, physiological and genomic analyses suggest that the three novel isolates represent two novel species of a novel genus in the family Planctomycetaceae. We thus propose to introduce the genus Symmachiella gen. nov. Strains Mal52T and Pan258 are assigned to the species Symmachiella dynata sp. nov. and CA54T to Symmachiella macrocystis sp. nov. Strains Mal52T and CA54T represent the respective type strains of the novel species.
Symmachiella gen. nov.
Symmachiella (Sym.ma.chi.el’la N.L. fem. n. Symmachiella dim. of Gr. symmachia a union, an alliance; bacteria that aggregate). Members of the genus have a cell envelope architecture resembling that of Gram-negative bacteria, are aerobic, neutrophilic, mesophilic and heterotrophic. Cells divide by polar budding and form strong aggregates. Species of the genus lack pigmentation. The DNA G + C content is around 55%. The genus is part of the family Planctomycetaceae, order Planctomycetales, class Planctomycetia, phylum Planctomycetes. The type species of the genus is Symmachiella dynata.
Symmachiella dynata sp. nov.
Symmachiella dynata (dy.na’ta. N.L. fem. adj. dynata of Gr. dynate strong, intense; corresponding to the strong cohesion between the cells). In addition to the genus characteristics, cells of the species are ovoid or pear-shaped. Cells of the type strain grow between 10 and 30 °C (optimum 24 °C) and at pH 5.0 to 9.5 (optimum pH 7.5). The DNA G + C content of the type strain is 55.3%. The type strain is Mal52T (DSM 101177T = VKM B-3432T), which was isolated from an alga close to the coast of S’Arenal on the island Mallorca, Spain. Strain Pan258 (DSM 103143 = VKM B-3436) is an additional member of the novel species.
Symmachiella macrocystis sp. nov.
Symmachiella macrocystis (ma.cro.cys’tis. N.L. gen. n. macrocystis of Macrocystis; corresponding to the isolation of the strain from the giant kelp Macrocystis pyrifera). In addition to the genus characteristics, the cell shape is not uniform and can range from ovoid to rod-shape. The type strain is CA54T (DSM 104301T = VKM B-3450T), isolated from the giant bladder kelp Macrocystis pyrifera in Monterey Bay, California, USA. Growth of the type strain was observed at a temperature range of 15–24 °C (optimum at 22 °C) and at pH 6.5–9.5 (optimum at pH 7.5). The DNA G + C content of the type strain is 55.2%.
References
Acehan D, Santarella-Mellwig R, Devos DP (2013) A bacterial tubulovesicular network. J Cell Sci 127::277–280
Bengtsson MM, Øvreås L (2010) Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol 10:261
Boedeker C, Schuler M, Reintjes G, Jeske O, van Teeseling MC, Jogler M, Rast P, Borchert D, Devos DP, Kucklick M, Schaffer M, Kolter R, van Niftrik L, Engelmann S, Amann R, Rohde M, Engelhardt H, Jogler C (2017) Determining the bacterial cell biology of Planctomycetes. Nat Commun 8:14853
Boersma AS, Kallscheuer N, Wiegand S, Rast P, Peeters SH, Mesman RJ, Heuer A, Boedeker C, Jetten MS, Rohde M, Jogler M, Jogler C (2019) Alienimonas californiensis gen. nov. sp. nov., a novel Planctomycete isolated from the kelp forest in Monterey Bay. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01367-4
Bondoso J, Harder J, Lage OM (2013) rpoB gene as a novel molecular marker to infer phylogeny in Planctomycetales. Antonie Van Leeuwenhoek 104:477–488
Bondoso J, Balague V, Gasol JM, Lage OM (2014) Community composition of the Planctomycetes associated with different macroalgae. FEMS Microbiol Ecol 88:445–456
Bondoso J, Albuquerque L, Nobre MF, Lobo-da-Cunha A, da Costa MS, Lage OM (2015) Roseimaritima ulvae gen. nov., sp. nov. and Rubripirellula obstinata gen. nov., sp. nov. two novel planctomycetes isolated from the epiphytic community of macroalgae. Syst Appl Microbiol 38:8–15
Bondoso J, Godoy-Vitorino F, Balague V, Gasol JM, Harder J, Lage OM (2017) Epiphytic Planctomycetes communities associated with three main groups of macroalgae. FEMS Microbiol Ecol 93:fiw255
Castresana J (2000) Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol Biol Evol 17::540–552
Dedysh SN, Henke P, Ivanova AA, Kulichevskaya IS, Philippov DA, Meier-Kolthoff JP, Göker M, Huang S, Overmann J (2020a) 100‐year‐old enigma solved: identification, genomic characterization and biogeography of the yet uncultured Planctomyces bekefii. Environ Microbiol 22:198–211
Dedysh SN, Kulichevskaya IS, Beletsky AV, Ivanova AA, Rijpstra WIC, Damsté JSS, Mardanov AV, Ravin NV (2020b) Lacipirellula parvula gen. nov., sp. nov., representing a lineage of planctomycetes widespread in low-oxygen habitats, description of the family Lacipirellulaceae fam. nov. and proposal of the orders Pirellulales ord. nov., Gemmatales ord. nov. and Isosphaerales ord. nov. Syst Appl Microbiol 43:126050
Devos DP (2014) Re-interpretation of the evidence for the PVC cell plan supports a Gram-negative origin. Antonie Van Leeuwenhoek 105:271–274
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 32:1792–1797
Frank O, Michael V, Pauker O, Boedeker C, Jogler C, Rohde M, Petersen J (2014) Plasmid curing and the loss of grip—the 65-kb replicon of Phaeobacter inhibens DSM 17395 is required for biofilm formation, motility and the colonization of marine algae. Syst Appl Microbiol 38:120–127
Fuerst JA, Webb RI (1991) Membrane-bounded nucleoid in the eubacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA 88:8184–8188
Gimesi N (1924) Hydrobiologiai talmanyok (Hydrobiologische Studien). I. Planktomyces bekefii Gim. nov. gen. et sp., Budapest, Kiadja a Magyar Ciszterci. Rend, 1924, pp. 1–8
Graça AP, Calisto R, Lage OM (2016) Planctomycetes as novel source of bioactive molecules. Front Microbiol 7:1241
Hirsch P (1972) Two identical genera of budding and stalked bacteria: Planctomyces Gimesi 1924 and Blastocaulis Henrici and Johnson 1935. Int J Syst Evol Microbiol 22:107–111
Ivanova AA, Naumoff DG, Miroshnikov KK, Liesack W, Dedysh SN (2017) Comparative genomics of four isosphaeraceae planctomycetes: a common pool of plasmids and glycoside hydrolase genes sharedby Paludisphaera borealis PX4T, Isosphaera pallida IS1BT, Singulisphaera acidiphila DSM 18658T, and strain SH-PL62. Front Microbiol 8:412
Jeske O, Jogler M, Petersen J, Sikorski J, Jogler C (2013) From genome mining to phenotypic microarrays: planctomycetes as source for novel bioactive molecules. Antonie Van Leeuwenhoek 104:551–567
Jeske O, Schuler M, Schumann P, Schneider A, Boedeker C, Jogler M, Bollschweiler D, Rohde M, Mayer C, Engelhardt H, Spring S, Jogler C (2015) Planctomycetes do possess a peptidoglycan cell wall. Nat Commun 6:7116
Jogler M, Jogler C (2013) Towards the development of genetic tools for Planctomycetes. In: Fuerst JA (ed) Planctomycetes: cell structure, origins and biology. Springer, Berlin, pp 141–164
Jogler C, Glöckner FO, Kolter R (2011) Characterization of Planctomyces limnophilus and development of genetic tools for its manipulation establish it as a model species for the phylum Planctomycetes. Appl Environ Microbiol 77:5826–5829
Jogler C, Waldmann J, Huang X, Jogler M, Glöckner FO, Mascher T, Kolter R (2012) Identification of proteins likely to be involved in morphogenesis, cell division, and signal transduction in Planctomycetes by comparative genomics. J Bacteriol 194:6419–6430
Kallscheuer N, Jogler M, Wiegand S, Peeters SH, Heuer A, Boedeker C, Jetten MS, Rohde M, Jogler C (2019a) Rubinisphaera italica sp. nov. isolated from a hydrothermal area in the Tyrrhenian Sea close to the volcanic island Panarea. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01329-w
Kallscheuer N, Wiegand S, Jogler M, Boedeker C, Peeters SH, Rast P, Heuer A, Jetten MS, Rohde M, Jogler C (2019b) Rhodopirellula heiligendammensis sp. nov., Rhodopirellula pilleata sp. nov., and Rhodopirellula solitaria sp. nov. isolated from natural or artificial marine surfaces in Northern Germany and California, USA, and emended description of the genus Rhodopirellula. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01366-5
Kallscheuer N, Wiegand S, Peeters SH, Jogler M, Boedeker C, Heuer A, Rast P, Jetten MS, Rohde M, Jogler C (2019c) Description of three bacterial strains belonging to the new genus Novipirellula gen. nov., reclassificiation of Rhodopirellula rosea and Rhodopirellula caenicola and readjustment of the genus threshold of the phylogenetic marker rpoB for Planctomycetaceae. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01374-5
Kallscheuer N, Wiegand S, Boedeker C, Peeters SH, Jogler M, Rast P, Heuer A, Jetten MS, Rohde M, Jogler C (2020a) Aureliella helgolandensis gen. nov., sp. nov., a novel Planctomycete isolated from a jellyfish at the shore of the island Helgoland. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-020-01403-8
Kallscheuer N, Wiegand S, Heuer A, Rensink S, Boersma AS, Jogler M, Boedeker C, Peeters SH, Rast P, Jetten MS, Rohde M, Jogler C (2020b) Blastopirellula retiformator sp. nov. isolated from the shallow-sea hydrothermal vent system close to Panarea Island. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01377-2
Kartal B, de Almeida NM, Maalcke WJ, Op den Camp HJ, Jetten MS, Keltjens JT (2013) How to make a living from anaerobic ammonium oxidation. FEMS Microbiol Rev 37::428–461
Kim M, Oh H-S, Park S-C, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Kohn T, Wiegand S, Boedeker C, Rast P, Heuer A, Jetten M, Schüler M, Becker S, Rohde C, Müller R-W, Brümmer F, Rohde M, Engelhardt H, Jogler M, Jogler C (2020) Planctopirus ephydatiae, a novel Planctomycete isolated from a freshwater sponge. Syst Appl Microbiol 43:126022
König E, Schlesner H, Hirsch P (1984) Cell wall studies on budding bacteria of the Planctomyces/Pasteuria group and on a Prosthecomicrobium sp. Arch Microbiol 138:200–205
Konstantinidis KT, Rodriguez-R LM (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints 4: e1900v1
Konstantinidis KT, Tiedje JM (2005) Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187:6258–6264
Lachnit T, Fischer M, Kunzel S, Baines JF, Harder T (2013) Compounds associated with algal surfaces mediate epiphytic colonization of the marine macroalga Fucus vesiculosus. FEMS Microbiol Ecol 84:411–420
Lage OM, Bondoso J (2014) Planctomycetes and macroalgae, a striking association. Front Microbiol 5:267
Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ (2011) Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinform 12:124
Lee I, Ouk Kim Y, Park S-C, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Lindsay MR, Webb RI, Fuerst JA (1997) Pirellulosomes: A new type of membrane-bounded cell compartment in planctomycete bacteria of the genus Pirellula. Microbiology 143:739–748
Lonhienne TG, Sagulenko E, Webb RI, Lee KC, Franke J, Devos DP, Nouwens A, Carroll BJ, Fuerst JA (2010) Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA 107:12883–12888
Peeters SH, Wiegand S, Kallscheuer N, Jogler M, Heuer A, Jetten MS, Rast P, Boedeker C, Rohde M, Jogler C (2020) Three marine strains constitute the novel genus and species Crateriforma conspicua in the phylum Planctomycetes. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01375-4
Pilhofer M, Rappl K, Eckl C, Bauer AP, Ludwig W, Schleifer KH, Petroni G (2008) Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J Bacteriol 190:3192–3202
Pruesse E, Peplies J, Glöckner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal. RNA genes Bioinformatics 28:1823–1829
Qin Q-L, Xie B-B, Zhang X-Y, Chen X-L, Zhou B-C, Zhou J, Oren A, Zhang Y-Z (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196:2210–2215
Rast P, Glöckner I, Boedeker C, Jeske O, Wiegand S, Reinhardt R, Schumann P, Rohde M, Spring S, Glöckner FO (2017) Three novel species with peptidoglycan cell walls form the new genus Lacunisphaera gen. nov. in the family Opitutaceae of the verrucomicrobial subdivision 4. Front Microbiol 8:202
Rivas-Marin E, Devos DP (2018) The Paradigms They Are a-Changin’: past, present and future of PVC bacteria research. Antonie Van Leeuwenhoek 111:785–799
Rivas-Marin E, Canosa I, Santero E, Devos DP (2016) Development of Genetic Tools for the Manipulation of the Planctomycetes. Front Microbiol 7:914
Rivas-Marin E, Wiegand S, Kallscheuer N, Jogler M, Peeters S, Heuer A, Jetten MS, Boedeker C, Rohde M, Devos DP, Jogler C (2020) Maioricimonas rarisocia gen. nov., sp. nov., isolated from marine sediments close to Mallorca Island. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-020-01436-z
Scheuner C, Tindall BJ, Lu M, Nolan M, Lapidus A, Cheng J-F, Goodwin L, Pitluck S, Huntemann M, Liolios K (2014) Complete genome sequence of Planctomyces brasiliensis type strain (DSM 5305T), phylogenomic analysis and reclassification of Planctomycetes including the descriptions of Gimesia gen. nov., Planctopirus gen. nov. and Rubinisphaera gen. nov. and emended descriptions of the order Planctomycetales and the family Planctomycetaceae. Stand Gen Sci 9:10
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Van Teeseling MC, Mesman RJ, Kuru E, Espaillat A, Cava F, Brun YV, VanNieuwenhze MS, Kartal B, Van Niftrik L (2015) Anammox Planctomycetes have a peptidoglycan cell wall. Nat Commun 6:6878
Wagner M, Horn M (2006) The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17:241–249
Wallner SR, Bauer M, Würdemann C, Wecker P, Glöckner FO, Faber K (2005) Highly enantioselective sec-alkyl sulfatase activity of the marine planctomycete Rhodopirellula baltica shows retention of configuration. Angew Chem Int Ed 44:6381–6384
Wegner CE, Richter-Heitmann T, Klindworth A, Klockow C, Richter M, Achstetter T, Glockner FO, Harder J (2013) Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar Genom 9:51–61
Wiegand S, Jogler M, Jogler C (2018) On the maverick Planctomycetes. FEMS Microbiol Rev 42:739–760
Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, Kohn T, Peeters SH, Heuer A, Rast P, Oberbeckmann S, Bunk B, Jeske O, Meyerdierks A, Storesund JE, Kallscheuer N, Lücker S, Lage OM, Pohl T, Merkel BJ, Hornburger P, Müller R-W, Brümmer F, Labrenz M, Spormann AM, Op den Camp HJM, Overmann J, Amann R, Jetten MSM, Mascher T, Medema MH, Devos DP, Kaster A-K, Øvreås L, Rohde M, Galperin MY, Jogler C (2020) Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat Microbiol 5:126–140
Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer K-H, Whitman WB, Euzéby J, Amann R, Rosselló-Móra R (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645
Acknowledgements
Part of this research was funded by the Deutsche Forschungsgemeinschaft Grants KA 4967/1–1 and JO 893/4 − 1, Grant ALWOP.308 of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO), SIAM (Soehngen Institute for Anaerobic Microbiology) Grant No. 024002002 and the Radboud Excellence fellowship. We thank Ina Schleicher for skillful technical assistance. We thank Brian Tindall and Regine Fähnrich from the DSMZ as well as the staff from the All-Russian Collection of Microorganisms for support during strain deposition. We thank Anne-Kristin Kaster (KIT Karlsruhe, Germany) and Alfred M. Spormann (Stanford, USA) as well as the Aquarius Dive Shop Monterey and the Hopkins Marine Station for sampling support. We also thank our collaborators Sonja Oberbeckmann and Matthias Labrenz (IOW Warnemünde, Germany) for sampling support.
Funding
Open Access funding provided by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
MS and MW wrote the manuscript, NK prepared the figures and contributed to text preparation, SW and MJ performed the genomic and phylogenetic analyses, AH and PR isolated the strains and performed the initial cultivation and strain deposition, SHP and CB performed the light microscopic analysis, MSMJ contributed to text preparation and revised the manuscript, MR performed the electron microscopic analysis, CJ took the samples, supervised PR and AH and the study. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Salbreiter, M., Waqqas, M., Jogler, M. et al. Three Planctomycetes isolated from biotic surfaces in the Mediterranean Sea and the Pacific Ocean constitute the novel species Symmachiella dynata gen. nov., sp. nov. and Symmachiella macrocystis sp. nov.. Antonie van Leeuwenhoek 113, 1965–1977 (2020). https://doi.org/10.1007/s10482-020-01464-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-020-01464-9