Abstract
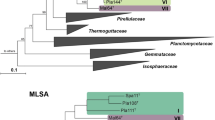
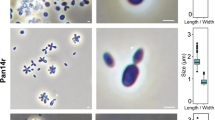
Access to axenic cultures of Planctomycetes is crucial for further investigating their complex lifestyle, uncommon cell biology and primary and secondary metabolism. As a contribution to achieve this goal in the future, we here describe three strains belonging to the novel genus Novipirellula gen. nov. The strains were isolated from biotic and abiotic surfaces in the Baltic Sea and from the island Heligoland in the North Sea. Colony colours range from white to light pink. Cells are acorn-shaped and grew optimally at neutral pH and temperatures between 27 and 30 °C. Phylogenetic analyses revealed that the isolated strains represent three novel species belonging to a new genus, Novipirellula gen. nov. Beyond that, our analysis suggests that Rhodopirellula rosea LHWP3T, Rhodopirellula caenicola YM26-125T and Rhodopirellula maiorica SM1 are also members of this novel genus. Splitting the current genus Rhodopirellula into a more strictly defined genus Rhodopirellula and Novipirellula also allowed readjusting the genus threshold value for the gene rpoB, encoding the RNA polymerase β-subunit, which is used as phylogenetic marker for Planctomycetales. A threshold range of 75.5–78% identity of the analysed partial rpoB sequence turned out to be reliable for differentiation of genera within the family Planctomycetaceae.





Similar content being viewed by others
References
Bengtsson MM, Sjøtun K, Lanzén A, Øvreås L (2012) Bacterial diversity in relation to secondary production and succession on surfaces of the kelp Laminaria hyperborea. ISME J 6:2188–2198
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucl Acids Res 47:W81–W87
Boedeker C, Schuler M, Reintjes G, Jeske O, van Teeseling MC, Jogler M, Rast P, Borchert D, Devos DP, Kucklick M, Schaffer M, Kolter R, van Niftrik L, Engelmann S, Amann R, Rohde M, Engelhardt H, Jogler C (2017) Determining the bacterial cell biology of Planctomycetes. Nat Commun 8:14853
Boersma A, Kallscheuer N, Wiegand S, Rast R, Peeters S, Mesman R, Heuer A, Boedeker C, Jetten M, Rohde M, Jogler M, Jogler C (2019) Alienimonas californiensis gen. nov. sp. nov., a novel Planctomycete isolated from the kelp forest in Monterey Bay. Antonie van Leeuwenhoek, https://doi.org/10.1007/s10482-019-01367-4
Bondoso J, Harder J, Lage OM (2013) rpoB gene as a novel molecular marker to infer phylogeny in Planctomycetales. Antonie Van Leeuwenhoek 104:477–488
Bondoso J, Balague V, Gasol JM, Lage OM (2014) Community composition of the Planctomycetes associated with different macroalgae. FEMS Microbiol Ecol 88:445–456
Bondoso J, Albuquerque L, Nobre MF, Lobo-da-Cunha A, da Costa MS, Lage OM (2015) Roseimaritima ulvae gen. nov., sp. nov. and Rubripirellula obstinata gen. nov., sp. nov. two novel planctomycetes isolated from the epiphytic community of macroalgae. Syst Appl Microbiol 38:8–15
Bondoso J, Godoy-Vitorino F, Balague V, Gasol JM, Harder J, Lage OM (2017) Epiphytic Planctomycetes communities associated with three main groups of macroalgae. FEMS Microbiol Ecol 93: fiw255
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Cayrou C, Raoult D, Drancourt M (2010) Broad-spectrum antibiotic resistance of Planctomycetes organisms determined by Etest. J Antimicrob Chemother 65:2119–2122
Devos DP (2014) Re-interpretation of the evidence for the PVC cell plan supports a Gram-negative origin. Antonie Van Leeuwenhoek 105:271–274
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 32:1792–1797
Frank CS (2011) Polyphasische Taxonomie, Kerngenom und Lebenszyklus von Rhodopirellula-Stämmen. Ph. D. thesis, University of Bremen, Germany
Fuerst JA, Sagulenko E (2011) Beyond the bacterium: Planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9:403–413
Godinho O, Calisto R, Ovreas L, Quinteira S, Lage OM (2019) Antibiotic susceptibility of marine Planctomycetes. Antonie Van Leeuwenhoek 112:1273–1280
Jeske O, Schüler M, Schumann P, Schneider A, Boedeker C, Jogler M, Bollschweiler D, Rohde M, Mayer C, Engelhardt H (2015) Planctomycetes do possess a peptidoglycan cell wall. Nat Commun 6:7116
Jogler M, Jogler C (2013) Towards the development of genetic tools for Planctomycetes. In: Fuerst JA (ed) Planctomycetes: cell structure, origins and biology. Springer, New York, pp 141–164
Jogler C, Glöckner FO, Kolter R (2011) Characterization of Planctomyces limnophilus and development of genetic tools for its manipulation establish it as a model species for the phylum Planctomycetes. Appl Environ Microbiol 77:5826–5829
Jogler C, Waldmann J, Huang X, Jogler M, Glöckner FO, Mascher T, Kolter R (2012) Identification of proteins likely to be involved in morphogenesis, cell division, and signal transduction in Planctomycetes by comparative genomics. J Bacteriol 194:6419–6430
Kallscheuer N, Jogler M, Wiegand S, Peeters S, Heuer A, Boedeker C, Jetten M, Rohde M, Jogler C (2019a) Rubinisphaera italica sp. nov. isolated from a hydrothermal area in the Tyrrhenian Sea close to the volcanic island Panarea. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01329-w
Kallscheuer N, Jogler M, Wiegand S, Peeters S, Heuer A, Boedeker C, Jetten M, Rohde M, Jogler C (2019b) Three novel Rubripirellula species isolated from artificial plastic surfaces submerged in the German part of the Baltic Sea and the estuary of the river Warnow. Antonie van Leeuwenhoek, https://doi.org/10.1007/s10482-019-01368-3
Kallscheuer N, Wiegand S, Jogler M, Boedeker C, Peeters S, Rast P, Heuer A, Jetten M, Rohde MCJ (2019c) Rhodopirellula heiligendammensis sp. nov., Rhodopirellula pilleata sp. nov., and Rhodopirellula solitaria sp. nov. isolated from natural or artificial marine surfaces in Northern Germany and California, USA. Antonie van Leeuwenhoek, https://doi.org/10.1007/s10482-019-01366-5
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Kohn T, Wiegand S, Boedeker C, Rast P, Heuer A, Jetten MSM, Schüler M, Becker S, Rohde C, Müller R-W, Brümmer F, Rohde M, Engelhardt H, Jogler M, Jogler C (2019) Planctopirus ephydatiae, a novel Planctomycete species isolated from the freshwater sponge Ephydatia fluviatilis. Syst Appl Microbiol. https://doi.org/10.1016/j.syapm.2019.126022
König E, Schlesner H, Hirsch P (1984) Cell wall studies on budding bacteria of the Planctomyces/Pasteuria group and on a Prosthecomicrobium sp. Arch Microbiol 138:200–205
Lage OM, Bondoso J (2014) Planctomycetes and macroalgae, a striking association. Front Microbiol 5:267
Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ (2011) Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinform 12:124
Lee I, Ouk Kim Y, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucl Acids Res 42:D490–D495
Lonhienne TG, Sagulenko E, Webb RI, Lee K-C, Franke J, Devos DP, Nouwens A, Carroll BJ, Fuerst JA (2010) Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA 107:12883–12888
Luo C, Rodriguez RL, Konstantinidis KT (2014) MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucl Acids Res 42:e73
Mitchell AL, Attwood TK, Babbitt PC et al (2019) InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucl Acids Res 47:D351–D360
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055
Peeters SH, van Niftrik L (2018) Trending topics and open questions in anaerobic ammonium oxidation. Curr Opin Chem Biol 49:45–52
Peeters S, Wiegand S, Kallscheuer N, Jogler M, Heuer A, Jetten M, Rast P, Boedeker C, Rohde M, Jogler C (2019) Three marine strains constitute the novel genus and species Crateriforma conspicua gen. nov. sp. nov. in the phylum Planctomycetes. Antonie van Leeuwenhoek, accepted manuscript (ANTO-D-19-00328)
Pilhofer M, Rappl K, Eckl C, Bauer AP, Ludwig W, Schleifer KH, Petroni G (2008) Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J Bacteriol 190:3192–3202
Pruesse E, Peplies J, Glöckner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829
Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, Zhou J, Oren A, Zhang YZ (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196:2210–2215
Rast P, Glöckner I, Boedeker C, Jeske O, Wiegand S, Reinhardt R, Schumann P, Rohde M, Spring S, Glöckner FO (2017) Three novel species with peptidoglycan cell walls form the new genus Lacunisphaera gen. nov. in the family Opitutaceae of the Verrucomicrobial subdivision 4. Front Microbiol 8:202
Richter M, Richter-Heitmann T, Klindworth A, Wegner C-E, Frank CS, Harder J, Glöckner FO (2014) Permanent draft genomes of the Rhodopirellula maiorica strain SM1. Mar Genomics 13:19–20
Rivas-Marin E, Canosa I, Santero E, Devos DP (2016) Development of genetic tools for the manipulation of the Planctomycetes. Front Microbiol 7:914
Rodriguez-R LM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ (preprints)
Roh SW, Lee H-W, Yim KJ, Shin N-R, Lee J, Whon TW, Lim N-L, Kim D, Bae J-W (2013) Rhodopirellula rosea sp. nov., a novel bacterium isolated from an ark clam Scapharca broughtonii. J Microbiol 51:301–304
Schlesner H (1986) Pirella marina sp. nov., a budding, peptidoglycan-less bacterium from brackish water. Syst Appl Microbiol 8:177–180
Schlesner H, Hirsch P (1984) Assignment of ATCC 27377 to Pirella gen. nov. as Pirella staleyi comb. nov. Int J Syst Evol Microbiol 34:492–495
Schlesner H, Hirsch P (1987) Rejection of the genus name Pirella for pear-shaped budding bacteria and proposal to create the genus Pirellula gen. nov. Int J Syst Evol Microbiol 37:441
Schlesner H, Rensmann C, Tindall BJ, Gade D, Rabus R, Pfeiffer S, Hirsch P (2004) Taxonomic heterogeneity within the Planctomycetales as derived by DNA–DNA hybridization, description of Rhodopirellula baltica gen. nov., sp. nov., transfer of Pirellula marina to the genus Blastopirellula gen. nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula. Int J Syst Evol Microbiol 54:1567–1580
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Spring S, Bunk B, Spröer C, Schumann P, Rohde M, Tindall BJ, Klenk H-P (2016) Characterization of the first cultured representative of Verrucomicrobia subdivision 5 indicates the proposal of a novel phylum. ISME J 10:2801
Stackebrandt E, Ebers J (2006) Taxonomic parameter revisited: tarnished gold standards. Microbiol Today 33:152–155
Staley JT (1973) Budding bacteria of the Pasteuria-Blastobacter group. Can J Microbiol 19:609–614
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Strous M, Fuerst JA, Kramer EH, Logemann S, Muyzer G, van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MS (1999) Missing lithotroph identified as new planctomycete. Nature 400:446–449
Van Teeseling MC, Mesman RJ, Kuru E, Espaillat A, Cava F, Brun YV, Van Nieuwenhze MS, Kartal B, Van Niftrik L (2015) Anammox Planctomycetes have a peptidoglycan cell wall. Nat Commun 6:6878
Vollmers J, Frentrup M, Rast P, Jogler C, Kaster AK (2017) Untangling genomes of novel planctomycetal and Verrucomicrobial species from monterey bay kelp forest metagenomes by refined binning. Front Microbiol 8:472
Wagner M, Horn M (2006) The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17:241–249
Ward N, Staley JT, Fuerst JA, Giovannoni S, Schlesner H, Stackebrandt E (2006) The order Planctomycetales, including the genera Planctomyces, Pirellula, Gemmata and Isosphaera and the Candidatus genera Brocadia, Kuenenia and Scalindua. Prokaryotes 7:757–793
Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, Kohn T, Peeters SH, Heuer A, Rast P, Oberbeckmann S, Bunk B, Jeske O, Meyerdierks A, Storesund JE, Kallscheuer N, Lücker S, Lage OM, Pohl T, Merkel BJ, Hornburger P, Müller R-W, Brümmer F, Labrenz M, Spormann AM, Op den Camp HJM, Overmann J, Amann R, Jetten MSM, Mascher T, Medema MH, Devos DP, Kaster A-K, Øvreås L, Rohde M, Galperin MY, Jogler C (2019) Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat Microbiol. https://doi.org/10.1038/s41564-019-0588-1
Winkelmann N, Harder J (2009) An improved isolation method for attached-living Planctomycetes of the genus Rhodopirellula. J Microbiol Meth 77:276–284
Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, Whitman WB, Euzeby J, Amann R, Rossello-Mora R (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645
Yoon J, Matsuo Y, Kasai H, Lee M-K (2014) Phylogenetic and taxonomic analyses of Rhodopirellula caenicola sp. nov., a new marine Planctomycetes species isolated from Iron Sand. J Phylogen Evol Biol 3:143
Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y (2018) dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucl Acids Res 46:W95–W101
Acknowledgements
Part of this research was funded by the Deutsche Forschungsgemeinschaft Grants KA 4967/1-1 and JO 893/4-1, Grant ALWOP.308 of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO), SIAM (Soehngen Institute for Anaerobic Microbiology) Grant no. 024002002 and the Radboud Excellence fellowship. We thank Ina Schleicher for skillful technical assistance. We also thank our collaborators Sonja Oberbeckmann, Matthias Labrenz (IOW Warnemünde, Germany), Jörn Petersen (DSMZ) and the Biological Institute Heligoland (BAH) for sampling support. We thank Brian Tindall and Regine Fähnrich as well as the BCCM/LMG Bacteria collection for on-going support during strain deposition.
Author information
Authors and Affiliations
Contributions
NK and SW wrote the manuscript, analysed data and prepared figures, AH, PR and MJ isolated the strains and performed the initial cultivation and strain deposition, SHP and CB performed the light microscopic analysis, MSMJ contributed to text preparation and revised the manuscript, MR performed the electron microscopic analysis, CJ, PR and AH took the samples in the Baltic Sea and on Heligoland, and CJ supervised the study. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kallscheuer, N., Wiegand, S., Peeters, S.H. et al. Description of three bacterial strains belonging to the new genus Novipirellula gen. nov., reclassificiation of Rhodopirellula rosea and Rhodopirellula caenicola and readjustment of the genus threshold of the phylogenetic marker rpoB for Planctomycetaceae. Antonie van Leeuwenhoek 113, 1779–1795 (2020). https://doi.org/10.1007/s10482-019-01374-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-019-01374-5