Abstract
Trifolium rubens L., commonly known as the red feather clover, is capable of symbiotic interactions with rhizobia. Up to now, no specific symbionts of T. rubens and their symbiotic compatibility with Trifolium spp. have been described. We characterized the genomic diversity of T. rubens symbionts by analyses of plasmid profiles and BOX–PCR. The phylogeny of T. rubens isolates was inferred based on the nucleotide sequences of 16S rRNA and two core genes (atpD, recA). The nodC phylogeny allowed classification of rhizobia nodulating T. rubens as Rhizobium leguminosarum symbiovar trifolii (Rlt). The symbiotic efficiency of the Rlt isolates was determined on four clover species: T. rubens, T. pratense, T. repens and T. resupinatum. We determined that Rlt strains formed mostly inefficient symbiosis with their native host plant T. rubens and weakly effective (sub-optimal) symbiosis with T. repens and T. pratense. The same Rlt strains were fully compatible in the symbiosis with T. resupinatum. T. rubens did not exhibit strict selectivity in regard to the symbionts and rhizobia closely related to Rhizobium grahamii, Rhizobium galegae and Agrobacterium radiobacter, which did not nodulate Trifolium spp., were found amongst T. rubens nodule isolates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In their symbiotic association with legume plants, rhizobia have the potential to fix nitrogen in amounts sufficient to reduce the dependence of plants on nitrogen fertilizers (Herridge 2008). They are distributed worldwide in many types of soil where they can be found as free-living organisms or symbionts of leguminous plants. Rhizobium-legume symbiosis plays a critical role in sustainable agriculture, because it reduces the need for nitrogen fertilizer while ensuring efficient protein-rich production. Rhizobia attach to the root hairs of plants, invade plant tissues, and colonize the cells, forming nodules where they differentiate into nitrogen-fixing bacteroids. The Rhizobium-legume symbiosis is specific and depends on the exchange of signal molecules, such as flavonoids, secreted by plants, which induce expression of bacterial nodulation (nod) genes via interaction with the NodD regulatory protein (Perret et al. 2000; Jones et al. 2007; Oldroyd and Downie 2008; Wang et al. 2012). The Nod proteins synthesize lipochitin oligosaccharides (Nod factors) recognized by host plant receptors and, in response, tubular structures called infection threads are formed where bacteria proliferate and are released into plant nodule cells forming symbiosomes. In these structures, bacteria differentiate into nitrogen-fixing bacteroids and turn N2 into ammonia, which is assimilated by the legume host (Heidstra and Bisseling 1996; Gage and Margolin 2000). In the indeterminate nodules formed by galegoid plants, five developmental zones are distinguished: the apical meristem functioning during nodule development (I), the invasion zone (II) into which infection threads release rhizobia, the interzone (II–III), the nitrogen-fixing zone (III), the senescence zone (IV), and the saprophytic zone (V) in older nodules (Vasse et al. 1990; Timmers et al. 2000). In the saprophytic zone, bacteroids degenerate and non-nitrogen fixing, undifferentiated rhizobia are released from infection threads, which increase the rhizobial population in the rhizosphere after nodule senescence (Timmers et al. 2000; Wielbo et al. 2010a, b). Thus, the nitrogen-fixing nodule is an organ where reciprocal benefits for both partners occur in different regions of the nodules. Although the legume-Rhizobium symbiosis is beneficial to the host, the nitrogen-fixation efficiency significantly varies between different plant-Rhizobium interactions and the molecular mechanisms of strain-specific nitrogen fixation are largely unknown (Schumpp and Deakin 2010; Wang et al. 2012). Plants play a significant role in the control of later stages of symbiosis, such as bacteroid differentiation inside nodules of some galegoid (IRLC) plants (Medicago, Trifolium, Vicia, Pisum, Astragalus) and endoreduplication of bacterial genomes forming bacteroids (Mergaert et al. 2006; Haag et al. 2013). At this stage, bacteroids exhibit decreased cytoplasmic membrane integrity and undergo terminal differentiation in relation to their free-living form while maintaining the metabolic activity required for nitrogen fixation and nutrient exchange with the host plant. Bacteroid differentiation studied in Medicago truncatula is mediated by a large family of legume nodule-specific cysteine-rich (NCR) peptides transported to symbiosomes, which have antimicrobial activity in vitro and have a critical role in bacteroid development and persistence in vivo (Haag et al. 2011; Van De Velde et al. 2010; Kondorosi et al. 2013).
The genome of Rhizobium leguminosarum is large and complex, consisting of a chromosome and a variable number of large plasmids (Young et al. 2006; Mazur et al. 2011; Kumar et al. 2015). Symbiotic functions are encoded by genes located in symbiotic plasmids (pSym) (Perret et al. 2000; Young et al. 2006; Mazur et al. 2011, 2013). The plasmids constitute a pool of accessory genetic information and contribute to the plasticity and dynamic state of the genome commonly observed among members of the Rhizobiaceae family (Palacios and Newton 2005). The host range of R. leguminosarum (Rl) species varies; R. leguminosarum bv. viciae (Rlv) is able to induce efficient symbiosis with legumes belonging to the genera Pisum, Vicia, Lathyrus, and Lens forming several species and biovars (symbiovars) (Laguerre et al. 2003; Alvarez-Martinez et al. 2009; Ramirez-Bahena et al. 2009; Rogel et al. 2011; Rashid et al. 2015). The development of effective symbiotic associations of Rlv with the large group of legume plants indicates that both partners contain compatible determinants for successful nodulation and N2 fixation. In contrast to Rlv, R. leguminosarum bv. trifolii (Rlt) symbiotic host range is confined to the clover genus (Trifolium spp.) (Ramirez-Bahena et al. 2008; Reeve et al. 2010a, b; Kumar et al. 2015).
The clover genus Trifolium L. is a member of a large clade of legumes lacking one copy of the chloroplast—inverted repeat (IRLC) and is one of the largest genera in the family consisting of approximately 255 herbaceous species (Watson and Sayed-Ahmed 2000; Ellison et al. 2006). These are commonly cultivated forage and green manure crops, which grow in a great range of soils with the highest species diversity in the temperate climate (Ellison et al. 2006). Rlt - Trifolium sp. symbiotic associations are mostly effective; however, host-dependent ineffective nodulation is evident amongst Rlt strains that nodulate clover species. Some Rlt strains form effective nodules on Trifolium repens, Trifolium pratense, and Trifolium subterraneum, whereas other strains are ineffective on Trifolium subterraneum (Tesfaye and Holl 1999). There are Rlt strains effective on T. subterraneum, but ineffective or partially effective on T. repens and T. pretense (Elliot et al. 1998). In another case, Rlt strains are able to form effective nodules on Trifolium ambiguum (Caucasian clover) but ineffective ones on T. repens, T. pratense and vice versa (Beauregard et al. 2004; Miller et al. 2007). Melino et al. (2012) reported sub-optimal or ineffective symbiotic association between an Rlt strain and T. subterraneum, T. purpureum, and Trifolium polymorphum. These reports show that rhizobial strains commonly found in agricultural soils are frequently poorly compatible with commercially grown clover species but little is known of the cause of their ineffectivity.
One of the clover species is Trifolium rubens L., commonly known as the red feather. This is a perennial, ornamental, xerothermic species of clover growing in normal soil in Central and South Europe. In Poland, T. rubens belongs to rare species whose occurrence has significantly decreased (86%) in the recent years (Michalik 2009). Like other clover species, it is capable of symbiotic association with rhizobia; however, to the best our knowledge, no specific symbionts of T. rubens species and their symbiotic efficiency have been described.
The primary goal of the study was to investigate the symbiotic compatibility of T. rubens nodule isolates with four agronomically important Trifolium spp. Moreover, genomic diversity and phylogeny of T. rubens symbionts were established. The symbiotic phylogeny inferred based on the nodC of R. leguminosarum nodulating T. rubens classified the isolates as belonging to the biovar trifolii. In clover plant tests, T. rubens isolates formed ineffective or weakly effective symbiosis with their native host. Their symbiotic efficiencies with other species i.e. T. repens, T. pratense and Trifolium resupinatum were also varied. T. rubens seems to not exhibit strict selectivity in regard to the symbionts and other rhizobia closely related to Rhizobium grahamii, Rhizobium galegae and Agrobacterium radiobacter, which were not able to nodulate four studied Trifolium spp., were found amongst R. leguminosarum bv. trifolii nodule isolates.
Materials and methods
Bacterial strains and growth conditions
Rhizobia were isolated from root nodules of T. rubens L. grown in meadows in the region of Lublin, Poland. The bacteria were isolated from surface-sterilized root nodules as follows: nodules were intensively washed in water and sterile distilled water and then nodules were sterilized by 3 min in 3% sodium chlorite and washed repeatedly in sterile distilled water. After surface sterilization, the nodules were crushed and their contents were plated on solid 79CA medium (Vincent 1970). The bacteria isolated from nodules were purified by repeatedly streaking of single colonies and pure cultures were used in further experiments.
DNA analyses
Plasmid content analyses of the nodule isolates were performed as described by Eckhardt (1978). Estimation of plasmids size was performed using Bio-Profile V11.01 (Vilber-Lourmat, France) compared with standard plasmids of R. leguminosarum bv. viciae strain 3841 (Young et al. 2006). Genomic DNA of each isolate was extracted from 5 ml of a 2-day bacterial culture in liquid TY using the method of Pitcher et al. (1989). PCR reactions were carried out using the Ready Mix Taq PCR Reaction (Sigma) according to the manufacturer’s recommendations. The primers and protocols used for amplification and sequencing the genes encoding 16S rRNA, atpD and recA (Weisburg et al. 1991; Louws et al. 1994; Gaunt et al. 2001; Martens et al. 2008) and for BOX-PCR are described in Table S1. Standard techniques were used for DNA labeling and Southern hybridization (Sambrook et al. 1989). DNA probe nodC for Southern hybridization was obtained by PCR amplification with R. leguminosarum bv. trifolii TA1 genomic DNA as the template and appropriate primers (Laguerre et al. 2001) (Table S1). The hybridization probe was labeled with the non-radioactive DIG DNA Labeling and Detection Kit (Roche). Automatic sequencing of PCR products was performed using the BigDye™ Terminator Cycle Sequencing Kit and an ABI Prism 3730 XL Genetic Analyzer (Applied Biosystems) according to manufacturer’s instructions. BOX-PCR genomic fingerprints were obtained as described by Louws et al. (1994). The sequences were aligned with those from GenBank using the MEGA5.05 software package (Tamura et al. 2011). The distances were calculated according to the Kimura’s 2-parameters (Kimura 1980). Phylogenetic trees were inferred using neighbor-joining method (NJ). Bootstrap analyses were calculated based on 1000 replications (Felsenstein 1985). Chromosomal 16S rRNA and the house-keeping genes atpD and recA, and symbiotic nodC sequences of T. rubens isolates determined in this study have been deposited in the GenBank under following accession numbers: KU715819-KU715834, KU714648-KU714656, KU714675-KU714689.
Plant growth experiments
To examine the symbiotic effectiveness of T. rubens isolates, four clover species were used as host plants: red feather (T. rubens L.), red clover (T. pratense L. cv. Rozeta), white clover (T. repens cv. Lipollo), and Persian clover (T. resupinatum L. Lightning). The seeds were surface sterilized by 15 min in 2% sodium hypochlorite, washed in sterile distilled water, and germinated on 0.8% agar-water plates. Four-day-old clover seedlings were planted in sterile nitrogen-free slants (one per tube) and 2 days later healthy seedlings were inoculated with ~108 cells of an individual strain grown in liquid 79CA, pelleted, and suspended in sterile water. Ten replicates were prepared in glass tubes for each rhizobial strain-clover species. The plants were grown for 5 weeks in a greenhouse under natural light supplemented with artificial light (14 h day/10 h night, at 24/19 °C). Then, the number of nodules and the fresh weight of shoots were examined. Symbiotic effectivity was estimated based on nodule number and the fresh shoot weight inoculated and uninoculated (growing without nitrogen) control plants.
A greenhouse experiment was carried out in plastic pots (500 cm3) filled with sterilized mixture of sand and vermiculite in the ratio 3:1. One day before seeding, each pot (two pots for each strain/clover species) was fertilized with 150 ml of Fahraeus N-free medium (negative control, N−) or the medium supplemented with 0.1% KNO3 (positive control, N+). Seeds of the four Trifolium spp. were surface sterilized for 15 min in 2% sodium hypochlorite, and then repeatedly washed with sterile distilled water. After sterilization, forty legume seeds per pot were seeded. After 5 days each pot was inoculated with 10 ml of rhizobia culture (~108 cfu ml−1). Plants were watered once a week with sterilized water and maintained under greenhouse conditions (day/night temperature: 23/18 °C; 12 h light/12 h dark period; relative humidity 70–80%). Plants were harvested 40 days after planting; wet mass and nodule number of thirty plants from each of the pots were estimated. The results of plant tests were analyzed with the Student’s test at a significance level of p < 0.05 and used for statistical analysis.
Confocal microscopy
Images of the root nodule sections were collected on a laser scanning confocal microscope LSM780 Zeiss with ZEN2010 data acquisition software using a EC Plan—Neofluar 10x/0.30 M27 objective. Sequential two-channel imaging in tile scan mode was performed with excitation light set at 488 nm from an Argon laser for SYTO9 and at 561 nm from a DPSS laser for Propidium Iodide (PI). Fluorescence emission was recorded in the range of 490–590 and 600–690 nm, respectively. Both lasers worked at 2% power to avoid photobleaching. Pinhole diameter was set to 1 AU. In situ live/dead staining of hand nodule sections was performed by incubation for 20 min in live/dead staining solution (5 μM SYTO9 and 30 μM PI in 50 mM Tris pH 7.0 buffer. Section were removed from the staining solution and mounted in deionized water for microscopy observation.
Results
Genomic diversity of T. rubens nodule isolates
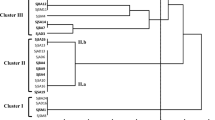
To examine the genomic diversity of T. rubens nodule isolates, DNA of 63 isolates were analyzed using the BOX-PCR analysis. The dendrogram constructed based on these results showed a high genomic diversity of the isolates, which were divided into two large groups at a low-level similarity (~1%) and further separated into numerous branches (Fig. 1). In total, 41 different DNA profiles with 23 characteristic for single isolates were found.
Plasmid patterns of the T. rubens nodule isolates
A group of 42 T. rubens nodule isolates representing different BOX-PCR profiles was selected and further investigated for the plasmid content using the Eckhardt lysis procedure (Fig. 2). In four isolates, plasmids were not identified. Other strains harbored from 1 to 6 plasmids with approximate molecular weight (m.w.) ranging from 70 to 1140 kb (data not shown). Isolates that differed in the plasmid patterns were assumed to be distinct strains. In a majority of the strains, a single symbiotic plasmid (pSym) with an approximate m.w. ranging from 263 to 458 kb was identified by Southern hybridization with the nodC probe derived from R. leguminosarum bv. trifolii TA1 (Mazur et al. 2011). pSym plasmids were not identified in 10 of the 42 strains. The total m.w. of the plasmids in the individual strains ranged from 1124 to 2885 kb with the average of 2176 kb, which constitutes 30.2% of the average genome size of R. leguminosarum bv. trifolii WSM2304 and WSM1325 of approx. 7.2 Mb (Reeve et al. 2010a, b). Table 1 shows the main symbiotic and genetic differences between the examined T. rubens isolates.
Phylogenetic analysis of T. rubens isolates
To determine the taxonomic status of T. rubens symbionts, fragments of 16S rDNA of 16 isolates were sequenced for genus identification. Phylogenetic analysis showed unambiguously a close relationship of these isolates to the type strains of Rhizobium spp. with 99.5–100% 16S rDNA sequence similarity (Fig. 3). Twelve of the 16 strains formed a separate, monophyletic group together with the Rhizobium spp. type strains with 100% similarity. Three strains were clearly distinct from the other isolates: strain Trb61 was closely related to Agrobacterium radiobacter CIP67.1T with 100% similarity and strain Trb30 was related to Neorhizobium galegae LMG6214T and R. vignae CCBAU05176T with pairwise sequence similarity 99.6 and 99.7%, respectively. Strain Trb142 was most similar (99.4%) to R. grahamii CCGE502T. For further species identification, the atpD and recA chromosomal genes of T. rubens isolates were sequenced. The phylogenetic trees constructed for the individual core genes using the neighbor-joining method (NJ) with high bootstrap replications showed that the strains which nodulate T. repens belong to the R. leguminosarum species (data not shown). Next, the concatenated sequences (759 bp) of two core genes (atpD, recA) were subjected to the NJ analyses, which allowed species identification of the isolates (Fig. 4). The topology of the phylogenetic tree was found to be very similar to that of the individual gene trees. The 6 isolates (Trb124, 107, 65, 116, 45, 75) forming the separate cluster were the most similar to R. leguminosarum bv. viciae 3841 (Young et al. 2006) (97.8–95.4%), R. leguminosarum bv. viciae USDA2370 (95.6–94.9%), R. leguminosarum bv. trifolii WSM1325 (Reeve et al. 2010b) and R. indigoferae CCBA71042T (95.1–94.3%) (Wu et al. 2011) showing their common taxonomic position (Table 2). The pairwise comparisons of the T. rubens isolates to the other relatives were 94.3–93.5% similar to R. pisi DSM 30132T, 93–94% to R. fabae CCBAU33202T, and 93–94% to R. leguminosarum bv. trifolii WSM2304. Strain Trb30 was most closely related to R. vignae CCBAU05176T and N. galegae LMG6214T with 97 and 96% similarity, respectively (Ren et al. 2011; Österman et al. 2015). Rhizobium sp. strain Trb142 was similar to Rhizobium sp. CCBAU15278 (92.5%) (Wu et al. 2011), but its taxonomic position remains unclear. The most distinct Trb61 was identified as A. radiobacter CIP67.1T with 99.5% similarity.
To examine the phylogenic history of symbiotic genes, the common nodC gene encoding N-acetylglucosaminyltransferase engaged in the first step of Nod factor synthesis was sequenced in the isolates identified as most similar to R. leguminosarum and the NJ phylogenetic tree was constructed (Fig. 5). The sampled T. rubens isolates formed a separate cluster with 97.3–100% sequence identity to one another. In this cluster, 4 isolates (Trb116, 124, 107, 75) possessed identical nodC, but some minor differences in nodC were found in strain Trb45, which showed 98.8% identity and in strain Trb65 with 95.8% identity with the other isolates. The nodC of 6 strains was highly similar to the nodC of R. leguminosarum bv. trifolii ATCC14480T, with sequence identity 96.7 and 93.8% in Trb65 (Table 2). Interestingly, the nodC sequences of T. rubens isolates were most similar (97.3 and 94.2% in Trb65) to nodC of the recently described new species Rhizobium aegyptiacum USDA 7124T isolated from nodules of Trifolium alexandrinum L., clover plants growing in different regions in Egypt and closely related to Rhizobium bangladeshense BLR175T isolated from lentil (Shamseldin et al. 2016; Rashid et al. 2015). Housekeeping chromosomal genes atpD and recA of R. aegyptiacum USDA 7124T were distantly related with 91.8–91.1% sequence similarity to the genes of T. rubens isolates, that confirms their belonging to different species and the same symbiovar trifolii. Other Rhizobium spp. strains form clearly separated symbiovars viciae and phaseoli (Fig. 5).
Based on the analysis of symbiotic phylogeny, 6 of the 9 sampled R. leguminosarum strains isolated from T. rubens nodules were unambiguously classified into biovar trifolii. We did not obtain nodC amplicons in the case of strains Trb30, 61, and 142, which is consistent with the negative result of Southern hybridization with the nodC probe in the plasmids assay (Fig. 2). Since these isolates were classified into the Rhizobium genus (Fig. 3), a significant polymorphism in nodC alleles or lack of nodC (e.g. Trb61) might be the causes of the lack of the nodC amplicons in these strains.
Host specificity and symbiotic efficiency of T. rubens nodule isolates
The selected T. rubens nodule isolates assigned to the genus Rhizobium (Fig. 3) were used for inoculation of clover plants in the tube experiments. Besides the red feather clover (T. rubens), the most commonly cultivated clovers were used: red clover (T. pratense L. cv. Rozeta), white clover (T. repens cv. Lipollo), and Persian clover (T. resupinatum L. Lightning). In the laboratory experiment, the clover plants were inoculated with 12 isolates and grown for 5 weeks. Only 8 strains classified as R. leguminosarum bv. trifolii (Table S2) formed nodules on the clovers. T. rubens nodulated by these strains formed 2.7–4.6 nodules per plant; the nodules were mostly small and white. The efficiency of nodulation measured as fresh weight of aerial clover parts was low, ranging from a 0.9 to 1.2-fold value (average 1.04) of fresh weight of shoots in relation to the uninoculated control. Only in the case of 3 (Trb5C.1, Trb65, Trb75) of the 8 isolates was the shoot weight difference significant, however, the symbiotic productivity was very low (Table S2). The other clovers i.e. T. pratense, T. repens, and T. resupinatum were nodulated by the same 8 rhizobial strains with various symbiotic effectiveness. In the case of T. pratense, 5 of the 8 strains formed low effective symbiosis (shoot weight average 1.8). In T. repens, 7 of the 8 strains showed increased fresh shoot weight (1.9 average), but the differences in shoot weight were significant only in 3 strains. The highest symbiotic productivity was recorded in Persian clover (T. resupinatum), and symbiotic efficiencies of 6 of the 8 strains were significant (2.6 average) relative to the uninoculated control. Strain Trb75, which inoculated four species of clover with a significant level of nitrogen fixation, was the most effective symbiont. Strains Trb116 and Trb78b induced the highest number of nodules on T. pratense and T. resupinatum; however, in both cases, the symbioses were inefficient and the values of the plant shoot weight were below those of the control plants.
In the pot plant assay, the most efficient T. rubens isolates (Trb45, Trb65, Trb75, Trb124) were selected for inoculation of four Trifolium spp. (Table 3; Fig. S1). The results confirmed the very low symbiotic efficiency of Rlt isolates in symbiosis with their native host T. rubens. Only in the case of Trb75 was the wet mass of aerial parts of clover over twice as much as that of the control. As in the tube assay, the most productive was the symbiosis of T. resupinatum with all the studied strains, yielding significant differences in wet aerial mass in all cases and from 3.9- to 13.6-fold higher wet mass. The symbiotic productivity of T. pratense was low and similar in four Rlt strains, yielding from 1.4- to 2.3-fold higher green plant mass. In the T. repens –Rlt association, the increase in the wet mass of plants inoculated with the studied strains was significant relative to control (fold 2.2–3.7); however, it was still lower than the T. resupinatum symbiotic productivity (Fig. S1). In conclusion, the efficiency of symbiosis of T. rubens isolates belonging to Rlt is host plant- and strain-dependent, indicating in most cases lack of symbiotic compatibility between rhizobia and T. rubens, T. pratense and T. repens host plants. The most compatible host plant for T. rubens isolates appeared to be T. resupinatum (Persian clover).
To study nodule organization, the 5-week-old nodules of T. rubens induced by effective (Trb75), low effective (Trb124), and ineffective (Trb65) strains were observed in confocal microscopy using a live/dead staining procedure involving application of a mixture of nucleic acid fluorescent dyes, SYTO09 and PI, to clover nodule sections (Fig. S2a, S2b). In the case of efficient nitrogen fixing nodules infected by Trb75, distinct developmental zones, i.e., a meristem zone (I), infection zone (II), and nitrogen fixation zone (III) were seen (Fig. S2b). The green fluorescent rhizobia filled the whole nitrogen-fixation zone, indicating that the bacteroids were live and metabolically active. However, in the same zone, numerous, red fluorescent dead bacteria were also observed (Fig. S2a). In the nodules infected by low effective Trb124, green live cells were clearly seen in the infection zone (II) and a few, green fluorescent live cells occupied the root-proximal oldest zone (IV); in the nitrogen-fixation zone, no live rhizobia were observed showing early abortion of bacteroid development. In the case of Trb65, which inefficiently infected T. rubens, only the infection zone with red fluorescent bacteria was seen; dead bacteria occupied the whole nodule indicating that the bacteria were rapidly killed and nodule development was arrested at the first stage of plant cell infection.
Besides the specific rhizobia infecting T. rubens, isolates that did not nodulate the clover species were found (Table S2). Among these, strain Trb30 was classified as most similar to R. galegae and R. vignae, strain Trb32 to R. phaseoli (data not shown), and Trb142 as Rhizobium sp. CCBAU15278 was most similar (89.6%) to R. grahamii CCGE502. Strain Trb61.2 was classified as A. radiobacter CIP67.1T (Fig. 3). Only in the case of Trb142-inoculated T. repens was some increase in plant growth observed (on average 1.6-fold, relative to the control) showing some plant promoting activity (Table S2).
Discussion
The present study revealed variable symbiotic responses of the host T. rubens, T. pratense, T. repens, and T. resupinatum clover species to inoculation with R. leguminosarum bv. trifolii isolated from T. rubens nodules. Both in the tube and pot plant assays, a majority of Rlt isolates from T. rubens nodules developed inefficient symbioses with their native host or weakly efficient with association with T. pratense and T. repens. The most productive, compatible symbiosis of T. rubens isolates was observed in the case of the agronomical important species T. resupinatum (Persian clover).
Several active nifHDKEN, nifB and fixA genes are required for Fix+ phenotype of Rlt (Perret et al. 2000). Commonly, Rlt strains isolated from red, white, or Persian clovers nodulated efficiently their original host plants (Beauregard et al. 2004). However, various responses of host plants from the same cross-inoculation group or the same species to a particular rhizobial strain have also been described, but the cause of this symbiotic incompatibility was mostly unexplained (Balatti and Pueppke 1992; Tesfaye and Holl 1999; Beauregard et al. 2004; Brito et al. 2008; Melino et al. 2012). Miller et al. (2007) described efficient symbiosis of the Rlt strain with T. ambiguum (Caucasian clover) and inefficient symbiosis with T. repens. In a molecular study of the nif/fix genes of this symbiont, the activity of the nifH-fixA intergenic region binding regulatory proteins was shown to be necessary for nifH expression which was responsible for host-specific efficient symbiosis of Rlt with T. ambiguum. Changes in the nucleotide sequence in the intergenic region affected the nifH expression resulting in the Fix− symbiotic phenotype (Miller et al. 2007). Other reports also suggested several regulatory mechanisms involved in host-specific symbiosis such as interaction of RpoN with the nifH promoter (Michiels et al. 1998). Cebolla et al. (1994) found that expression of Ensifer meliloti nifH and fixA promoters was very low in heterologous rhizobial backgrounds in comparison to homologous ones, which may be essential for host specificity. This effect was most strongly seen in the Rlt background. These observations show that optimal symbiotic nitrogen-fixation depends on mostly unknown plant and microbial regulatory factors.
Recently, a specific role of the NCR (NCR169) peptide in the differentiation and persistence of nitrogen fixing bacteroids in Medicago truncatula has been documented (Horváth et al. 2015; Price et al. 2015). Lack of this peptide in a M. truncatula mutant caused ineffective symbiosis with the specific symbiont. In the plant mutant, the bacterial differentiation was impaired and early senescence of symbiotic cells occurred. Complementation of mutation was associated with restoration of N2 fixation (Horváth et al. 2015). Price et al. (2015) found that the strain-host dependent symbiotic inefficiency in M. truncatula—E. meliloti interaction was dependent on specific degradation of NCR169 by a bacterial enzyme, zinc-metallopeptidase, encoded by gene hrrP located on the accessory plasmid. Lack of NCR169 affected the symbiotic outcome in late nodule development by triggering premature degeneration of differentiated nitrogen-fixing bacteroids and caused nodule senescence. The plasmid elimination or mutation in the hrrP gene caused the Fix+ phenotype of E. meliloti (Price et al. 2015). A critical role for symbiosis is also played by the BacA protein, which protects E. meliloti against the anti-bacterial action of NCR peptides allowing bacterial persistence within the nodule (Haag et al. 2011).
Since the symbionts of T. rubens have not been genetically described so far, the analyses of the genomic diversity and symbiotic phylogeny of the nodule isolates were performed. The pangenome of Rlt (Medini et al. 2005), consists of several strains with core genes shared by all the strains and a set of genes influencing the phenotypes and symbiotic activity of the strains (Kumar et al. 2015). A characteristic feature of the species is that it harbors several plasmids comprising a considerable number of accessory genes, which supports the broad genetic and phenotypic diversity of strains and species. The plasmids play a crucial role in rhizobial adaptation to specific environmental niches and strain evolution (Palacios and Newton 2005; Mazur and Koper 2012). The symbiotic genes of R. leguminosarum located on the pSym plasmid were also found to be highly variable and affected efficient symbioses with several host plants (Ramirez-Bahena et al. 2008, 2009; Marek-Kozaczuk et al. 2013). In this study, the genomes of the T. rubens nodule isolates were found to be highly differentiated based on the plasmid content showing considerable genetic variability as described earlier for Rlt isolated from nodules of T. pratense (Mazur et al. 2011, 2013). The taxonomic position of the selected T. rubens isolates was unambiguously defined as R. leguminosarum. The symbiotic phylogeny based of the nodC genes allowed defining the symbiovar of 6 isolates as Rlt that nodulated T. rubens, T. pratense, T. repens, and T. resupinatum. Fluorescence microscopy of Fix+ T. rubens nodule sections (Trb75) showed proper zonation; several plant cells were occupied by metabolically active bacteroids showing correct nodule development. The Fix+/Fix− T. rubens nodule section (Trb124) showed that the development of rhizobial infection was arrested in the infection zone and only a few plant cells in the oldest zone contained viable bacteroids reflecting residual nitrogen fixation observed in plant tests. In the Fix− T. rubens nodule sections (Trb65), the infection was also aborted in the infection zone but a vast majority of rhizobia were dead, corroborating inefficient symbiosis. Altogether, the morphological changes in nodule development confirmed the variable symbiotic activity in the interaction of the Rlt strains with the native clover T. rubens. We can speculate that some bacterial or plant molecular factors taking part in the later stages of nodule development, such as bacteroid terminal differentiation to nitrogen fixing forms are responsible for the different responses of the host plant to specific symbionts. These results show that the specificity in the clover symbiosis may not be limited to nodule formation, but might also be controlled at later stages of nodule development (Tesfaye and Holl 1999; Wielbo et al. 2010a; Price et al. 2015). Currently, the role of NCR plant peptides in the Trifolium - R. leguminosarum interactions has not been explored, but it is tempting to consider the involvement of the plasmid located genes in affecting the symbiotic phenotype of some host-rhizobia associations, especially in Rlt strains harboring several accessory plasmids. The mechanism by which a majority of Rlt isolates from T. rubens nodules do not fix nitrogen in symbiosis with their native T. rubens host is currently not understood.
The specific compatible rhizobia play the main role in plant growth, which is a consequence of nitrogen fixation. The legume root nodules are a specific environmental niche induced by competent rhizobia; however, multiple parasitic, symbiotic or endophytic species of bacteria can co-exist inside nodules despite the high selectivity of legume plants towards microsymbionts (Saidi et al. 2014; Sessitch et al. 2012). Endophytes are able to colonize plant internal tissues with minimal or no host damage (Schumpp and Deakin 2010; Zgadzaj et al. 2015). In this work, besides the Rlt strains isolated from the T. rubens nodules, we found strains that were highly similar to other species such as Trb61—A. radiobacter CIP67.1, Trb30—R. vignae, Trb142—R. grahamii CCGE 502, and several others with an undetermined taxonomic status. These rhizobia can populate nodules due to the reduced selectivity of T. rubens host plants towards symbionts. Poorly efficient symbiosis with T. rubens can favor non-specific infection, and symbionts and other bacteria coexisting in the nodules form some kind of facultative mutualism. These results are consistent with the hypothesis that the specificity in the clover symbiosis may not be limited to nodule establishment but might also be reflected at later stages of nodulation and/or development of effective nitrogen fixation (Schumpp and Deakin 2010).
References
Alvarez-Martinez ER, Valverde A, Ramirez-Bahena MH et al (2009) The analysis of core and symbiotic genes of rhizobia nodulating Vicia from different continents reveals their common phylogenetic origin and suggests the distribution of Rhizobium leguminosarum strains together with Vicia seeds. Arch Microbiol 191:659–668
Balatti PA, Pueppke SG (1992) Identification of North American soybean lines forming nitrogen-fixing nodules with Rhizobium fredi USDA257. Can J Plant Sci 72:49–55
Beauregard MS, Zheng W, Segui P (2004) Diversity of Trifolium ambiguum—nodulating rhizobia from the lower Caucasus. Biol Fertil Soils 40:128–135
Brito B, Toffanin A, Prieto RI et al (2008) Host-dependent expression of Rhizobium leguminosarum bv. viciae hydrogenase is controlled at transcriptional and post-transcriptional level. Mol Plant Microbe Interact 21:597–604
Cebolla A, Ruiz-Berraquero F, Palomares AJ (1994) Analysis of the expression from Rhizobium meliloti fix-promoters in other Rhizobium backgrounds. Microbiology 140:443–453
Eckhardt T (1978) A rapid method for the identification of plasmid deoxyribonucleic acid in bacteria. Plasmid 1:584–588
Elliot RM, Lowther WL, Ronson CW (1998) Interactions between and Trifolium repens in the field. In: Elmerich C, Kondorosi A, Newton WE (eds) Biological nitrogen fixation for the 21st century. Kluwer, Dordrecht
Ellison NW, Liston A, Steiner JJ et al (2006) Molecular phylogenetics of the clover genus (Trifolium – Leguminosae). Mol Phylogenet Evol 39:688–705
Felsenstein J (1985) Phylogenies and the comparative method. Am Nat 125:1–15
Gage DJ, Margolin W (2000) Hanging by a thread: invasion of legume plants by rhizobia. Curr Opin Microbiol 3:613–617
Gaunt MW, Turner SL, Rigottier-Gois L et al (2001) Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int J Syst Evol Microbiol 51:2037–2048
Haag AF, Sani M, Kerscher B et al (2011) Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol 9:e1001169
Haag AF, Arnold MF, Myka KK et al (2013) Molecular insights into bacteroid development during the Rhizobium-legume symbiosis. FEMS Microbiol Rev 37(3):364–383
Heidstra R, Bisseling T (1996) Nod factor-induced host responses and mechanisms of Nod factor perception. New Phytol 133:25–43
Herridge DF, Peoples MB, Boddey RM (2008) Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311:1–18
Horváth B, Domonkos Á, Kereszt A et al (2015) Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc Natl Acad Sci USA 112:3–8. doi:10.1073/pnas.1500777112
Jones KM, Kobayashi H, Davies BW et al (2007) How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5:619–633
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kondorosi E, Mergaert P, Kereszt A (2013) A paradigm for endosymbiotic life: cell differentiation of rhizobium bacteria provoked by host plant factors. Annu Rev Microbiol 67:611–628
Kumar N, Lad G, Giuntini E et al (2015) Bacterial genospecies that are not ecologically coherent: population genomics of Rhizobium leguminosarum. Open Biol 5:140133
Laguerre G, Nour SM, Macheret V et al (2001) Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 147:981–993
Laguerre G, Louvrier P, Allard MR et al (2003) Compatibility of rhizobial genotypes within natural populations of Rhizobium leguminosarum biovar viciae for nodulation of host legumes. Appl Environ Microbiol 69:2276–2283
Louws FJ, Fulbright DW, Stephens CT (1994) Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Environ Microbiol 60:2286–2295
Marek-Kozaczuk M, Leszcz A, Wielbo J et al (2013) Rhizobium pisi sv. trifolii K3.22 harboring nod genes of the Rhizobium leguminosarum sv. trifolii cluster. Syst Appl Microbiol 36(4):252–258
Martens M, Dawyndt P, Coopman R et al (2008) Advantages of for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int J Syst Evol Microbiol 58:200–214
Mazur A, Koper P (2012) Rhizobial plasmids—replication, structure and biological role. Cent Eur J Biol 7:571–586
Mazur A, Stasiak G, Wielbo J et al (2011) Intragenomic diversity of Rhizobium leguminosarum bv. trifolii clover nodule isolates. BMC Microbiol 11:123
Mazur A, Stasiak G, Wielbo J et al (2013) Phenotypic profiling of Rhizobium leguminosarum bv. trifolii clover nodule isolates reveal their both versatile and specialized metabolic capabilities. Arch Microbiol 195:255–267
Medini D, Donati C, Tettelin H et al (2005) The microbial pan-genome. Curr Opin Genet 15:589–594
Melino VJ, Drew E, Ballard R et al (2012) Identifying abnormalities in symbiotic development between Trifolium spp. and Rhizobium leguminosarum bv. trifolii leading to sub-optimal and ineffective nodule phenotypes. Ann Bot 206:1–14
Mergaert P, Uchiumi T, Alunni B et al (2006) Eucaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc Natl Acad Sci USA 103:5230–5235
Michalik S (2009) Changes in the number and distribution of selected xerodermic and mountain species in the permanent study plot “Czyżówki” in the years 1988–2007. Prądnik. Prace Muz Szafera 19:243–256
Michiels J, Moris M, Dombrecht B et al (1998) Differential regulation of Rhizobium etli rpoN2 gene expression during symbiosis and free-living growth. J Bacteriol 180(4):3620–3628
Miller SH, Elliot RM, Sullivan JT et al (2007) Host-specific regulation of symbiotic nitrogen fixation in Rhizobium leguminosarum biovar trifolii. Microbiology 153:3184–3195
Oldroyd GED, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59:519–546
Österman J, Mousavi SA, Koskinen P et al (2015) Genomic features separating ten strains of Neorhizobium galegae with different symbiotic phenotypes. BMC Genomics 16:348
Palacios R, Newton WE (eds) (2005) Genomes and genomics of nitrogen-fixing organisms. Springer, Dordrecht
Perret X, Staehelin C, Broughton WJ (2000) Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev 64:180–201
Pitcher DG, Saunders NA, Owen RJ (1989) Rapid extraction of bacterial genomic DNA with guanidine thiocyanate. Lett Appl Microbiol 8:151–156
Price PA, Tanner HR, Dillon BA et al (2015) Rhizobial peptidase HrrP cleaves host-encoded signaling peptides and mediates symbiotic compatibility. Proc Natl Acad Sci USA 112:15244–15249
Ramirez-Bahena MH, Garcia-Fraile P, Peix A et al (2008) Revision of the taxonomic status of the species Rhizobium leguminosarum (Frank 1879) Frank 1889AL. Rhizobium phaseoli Dangeard 1926AL and Rhizobium trifolii Dangeard 1926AL. R. trifolii is a later synonym of R. leguminosarum. Reclassification of the strain R. leguminosarum DSM 30132 (=NCIMB 11478) as Rhizobium pisi sp. nov. Int J Syst Evol Microbiol 58:2484–2490
Ramirez-Bahena MH, Velazquez E, Fernandez-Santos F et al (2009) Phenotypic, genotypic, and symbiotic diversities in strains nodulating clover in different soils in Spain. Can J Microbiol 55:1207–1216
Rashid MH, Young JP, Everall I, Clercx P et al (2015) Average nucleotide identity of genome sequences supports the description of Rhizobium lentis sp. nov., Rhizobium bangladeshense sp. nov. and Rhizobium binae sp. nov. from lentil (Lens culinaris) nodules. Int J Syst Evol Microbiol 65:3037–3045
Reeve W, O’Hara G, Chain P et al (2010a) Complete genome sequence of Rhizobium leguminosarum bv. trifolii strain WSM2304, an effective microsymbiont of the South American clover Trifolium polymorphum. Stand Genom Sci 2:66–76
Reeve W, O’Hara G, Chain P et al (2010b) Complete genome sequence of Rhizobium leguminosarum bv. trifolii strain WSM1325, an effective microsymbiont of annual Mediterranean clovers. Stand Genom Sci 2:347–356
Ren DW, Chen WF, Sui XH et al (2011) Rhizobium vignae sp. nov., a symbiotic bacterium isolated from multiple legume species. Int J Syst Evol Microbiol 61:580–586
Rogel MA, Ormeno-Orrillo E, Martinez-Romero E (2011) Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst Appl Microbiol 34:96–104
Saidi S, Ramirez-Bahena MH, Santillana N et al (2014) Rhizobium laguerreae sp. nov., nodulates Vicia faba on several continents. Int J Syst Evol Microbiol 64:242–247
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schumpp O, Deakin WJ (2010) How inefficient rhizobia prolong their existence within nodules. Trends Plant Sci 15:189–195
Sessitch A, Hardoim P, Döring J et al (2012) Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol Plant-Microbe Interact 25:28–36
Shamseldin A, Carro L, Peix A et al (2016) The symbiovar trifolii of Rhizobium bangladeshense and Rhizobium aegyptiacum sp. nov. nodulate Trifolium alexandrinum in Egypt. Syst Appl Microbiol 39(4):275–279
Tamura K, Peterson D, Peterson N et al (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tesfaye M, Holl FB (1999) Rhizobium strains that nodulate Trifolium semipilosum Fres. are phylogenetically distinct. Plant Soil 207:147–154
Timmers AC, Soupène E, Auriac MC et al (2000) Saprophytic intracellular rhizobia in alfalfa nodules. Mol Plant Microbe Interact 13:1204–1213
Van de Velde W, Zehirov G, Szatmari A et al (2010) Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327:1122–1126. doi:10.1126/science.1184057
Vasse J, de Billy F, Camut S et al (1990) Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J Bacteriol 172:4295–4306
Vincent JM (1970) A manual for the practical study of root nodule bacteria. International biological program handbook no. 15, Blackwell Scientific Publications Ltd, Oxford
Wang D, Yang S, Tang F et al (2012) Symbiosi specificity in the legume—rhizobial mutualism. Cell Microbiol 14:334–342
Watson LE, Sayed-Ahmed H, Badr A (2000) Molecular phylogeny of Old World Trifolium (Fabaceae), based on plastid and nuclear markers. Plant Syst Evol 224:153–171
Weisburg WG, Barns SM, Pelletier DA et al (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Wielbo J, Kuske J, Marek-Kozaczuk M et al (2010a) The competition between Rhizobium leguminosarum bv. viciae strains progresses until late stages of symbiosis. Plant Soil 337:125–135
Wielbo J, Marek-Kozaczuk M, Mazur A et al (2010b) Genetic and metabolic divergence within a Rhizobium leguminosarum bv. trifolii population recovered from clover nodules. Appl Environ Microbiol 76:4593–4600
Wu LJ, Wang HQ, Wang ET et al (2011) Genetic diversity of nodulating and non-nodulating rhizobia associated with soybean (Glycine soja Sieb. & Zucc.) in different ecoregions of China. FEMS Microbiol Ecol 76:439–450
Young JP, Crossman LC, Johnston AW et al (2006) The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol 7:R34
Zgadzaj R, James EK, Kelly S et al (2015) A legume genetic framework controls infection of nodules by symbiotic and endophytic bacteria. PLoS Genet 11:e1005280
Acknowledgements
The study was conducted by the research fund of Faculty of Biotechnology and Microbiology, Maria Curie-Skłodowska University, Lublin, Poland.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no financial and other conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10482_2017_922_MOESM1_ESM.tif
Fig. S1 Symbiotic performance (fresh soot weight and number of nodules per plant) of rhizobial strains Trb45, Trb65, Trb75, Trb124 with (a) Trifolium rubens, (b) T. pretense var. Rozeta, (c) T. repens var. Lipollo, (d) T. resupinatum var. Ligthning. +N uninoculated plant in medium supplemented with N source. −N negative control (uninoculated plant). Average values with standard deviations are shown. Asterisks indicate statistical significant differences (P value < 0.05). Supplementary material 1 (TIFF 511 kb)
10482_2017_922_MOESM2_ESM.tif
Fig. S2a. Confocal microscopy of nodule sections of Trifolium rubens inoculated with R. leguminosarum bv. trifolii Trb65, Trb75, Trb124 isolates presenting different symbiotic clover response. Nodules were stained with a mixture of SYTO9 (green signal) and PI (red). Live bacteria are stained by SYTO9 and dead bacteria with PI; co-localization green and red (merge). Supplementary material 2 (TIFF 412 kb)
10482_2017_922_MOESM3_ESM.tif
Fig. S2b. Confocal microscopy of nodule section of Trifolium rubens inoculated with R. leguminosarum bv. trifolii Trb75. On the left, developmental zones of nodule (I–IV) were marked. Supplementary material 3 (TIFF 790 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Marek-Kozaczuk, M., Wdowiak-Wróbel, S., Kalita, M. et al. Host-dependent symbiotic efficiency of Rhizobium leguminosarum bv. trifolii strains isolated from nodules of Trifolium rubens . Antonie van Leeuwenhoek 110, 1729–1744 (2017). https://doi.org/10.1007/s10482-017-0922-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-017-0922-7