Abstract
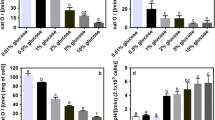
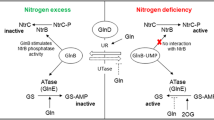
In the present work we studied the expression of genes from nitrogen central metabolism in the yeast Dekkera bruxellensis and under regulation by the Nitrogen Catabolite Repression mechanism (NCR). These analyses could shed some light on the biological mechanisms involved in the adaptation and survival of this yeast in the sugarcane fermentation process for ethanol production. Nitrogen sources (N-sources) in the form of ammonium, nitrate, glutamate or glutamine were investigated with or without the addition of methionine sulfoximine, which inhibits the activity of the enzyme glutamine synthetase and releases cells from NCR. The results showed that glutamine might act as an intracellular sensor for nitrogen availability in D. bruxellensis, by activating NCR. Gene expression analyses indicated the existence of two different GATA-dependent NCR pathways, identified as glutamine-dependent and glutamine-independent mechanisms. Moreover, nitrate is sensed as a non-preferential N-source and releases NCR to its higher level. After grouping genes according to their regulation pattern, we showed that genes for ammonium assimilation represent a regulon with almost constitutive expression, while permease encoding genes are mostly affected by the nitrogen sensor mechanism. On the other hand, nitrate assimilation genes constitute a regulon that is primarily subjected to induction by nitrate and, to a lesser extent, to a repressive mechanism by preferential N-sources. This observation explains our previous reports showing that nitrate is co-consumed with ammonium, a trait that enables D. bruxellensis cells to scavenge limiting N-sources in the industrial substrate and, therefore, to compete with Saccharomyces cerevisiae in this environment






Similar content being viewed by others
References
Basílio ACM, Araújo PRL, Morais JOF, da Silva Filho EA, de Morais Jr MA, Simões DA (2008) Detection and identification of wild yeast contaminants of the industrial fuel ethanol fermentation process. Curr Microbiol 5:322–326
Blomqvist J, Nogué VS, Gorwa-Grauslund M, Passoth V (2012) Physiological requirements for growth and competitiveness of Dekkera bruxellensis under oxygen-limited or anaerobic conditions. Yeast 29:265–274
Crauwels S, Van Assche A, de Jonge R, Borneman AR, Verreth C, Troels P, De Samblanx G, Marchal K, Van de Peer Y, Willems KA, Verstrepen KJ, Curtin CD, Lievens B (2015) Comparative phenomics and targeted use of genomics reveals variation in carbon and nitrogen assimilation among different Brettanomyces bruxellensis strains. Appl Microbiol Biotechnol 99:9123–9134
Crespo JL, Hall MN (2002) Elucidating TOR signalling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol Mol Biol Rev 66:579
Da Silva TC, Leite FC, De Morais Jr MA (2016) Distribution of Dekkera bruxellensis in a sugarcane-based fuel-ethanol fermentation plant. Lett Appl Microbiol 62:354–358
De Barros Pita W, Leite FCB, de Souza Liberal AT, Simões DA, de Morais Jr MA (2011) The ability to use nitrate confers advantage to Dekkera bruxellensis over S cerevisiae and can explain its adaptation to industrial fermentation processes. Antonie Van Leeuwenhoek 100:99–107
De Barros Pita W, Leite FCB, de Souza Liberal AT, Pereira LF, Carazzolle MF, Pereira GA, De Morais Jr MA (2012) A new set of reference genes for RT-qPCR assays in the yeast Dekkera bruxellensis. Can J Microbiol 58:1362–1367
De Barros Pita W, Castro Parente D, Simões DA, Passoth V, de Morais Jr MA (2013a) Physiology and gene expression profiles of Dekkera bruxellensis in response to carbon and nitrogen availability. Antonie Van Leeuwenhoek 104:855–868
De Barros Pita W, Tiukova I, Leite FC, Passoth V, Simões DA, de Morais Jr MA (2013b) The influence of nitrate on the physiology of the yeast Dekkera bruxellensis grown under oxygen limitation. Yeast 30:111–117
De Souza Liberal AT, Basílio ACM, Brasileiro BTRV, da Silva Filho EA, Simões DA, de Morais Jr MA (2007) Identification of the yeast Dekkera bruxellensis as major contaminant in continuous fuel ethanol fermentation. J Appl Microbiol 102:538–547
Flamholz A, Noor E, Bar-Even A, Milo R (2012) eQuilibrator—the biochemical thermodynamics calculator. Nucl Acids Res 40:D770–775
Galafassi S, Capusoni C, Moktaduzzaman M, Compagno C (2013) Utilization of nitrate abolishes the “Custers effect” in Dekkera bruxellensis and determines a different pattern of fermentation products. J Ind Microbiol Biotechnol 40:297–303
Garcia-Vallve S, Palau J, Romeu A (1999) Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol Biol Evol 16:1125–1134
Georis I, Tate JJ, Cooper TG, Dubois E (2011) Nitrogen-responsive regulation of GATA protein family activators Gln3 and Gat1 occurs by two distinct pathways, one inhibited by rapamycin and the other by methionine sulfoximine. J Biol Chem 286:44897–44912
Leite FCB, Basso TO, De Barros Pita WB, Gombert A, Simões DA, de Morais Jr MA (2012) Quantitative aerobic physiology of the yeast Dekkera bruxellensis, a major contaminant in bioethanol production plants. FEMS Yeast Res 13:34–43
Magasanik B, Kaiser CA (2002) Nitrogen regulation in Saccharomyces cerevisiae. Gene 290:1–18
Meimoun A, Holtzman T, Weissman Z, McBride HJ, Stillman DJ, Fink GR, Kornitzer D (2000) Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF(CDC4) ubiquitin-ligase complex. Mol Biol Cell 11:915–927
Piskur J, Ling Z, Marcet-Houben M, Ishchuk OP, Aerts A, LaButti K, Copeland A, Lindquist E, Barry K, Compagno C, Bisson L, Grigoriev IV, Gabaldon T, Phister T (2012) The genome of wine yeast Dekkera bruxellensis provides a tool to explore its food-related properties. Int J Food Microbiol 157:202–209
Riego L, Avendano A, DeLuna A, Rodriguez E, Gonzalez A (2002) GDH1 expression is regulated by GLN3, GCN4, and HAP4 under respiratory growth. Biochem Biophys Res Commun 293:79–85
Siverio JM (2002) Assimilation of nitrate by yeasts. FEMS Microbiol Rev 26:277–284
Teixeira MC, Monteiro PT, Guerreiro JF, Gonçalves JP, Mira NP, dos Santos SC, Cabrito TR, Palma M, Costa C, Francisco AP, Madeira SC, Oliveira AL, Freitas AT, Sá-Correia I (2014) The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucl Acids Res 42:D161–D166
ter Schure EG, van Rielb NAW, Verripsa CT (2000) The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol Rev 24:67–83
Walford SN (1996) Composition of Cane Juice. Proc S Afr Sug Technol Ass 70:265–266
Funding
This work was sponsored by the Bioethanol Research Network of the State of Pernambuco (CNPq-FACEPE/PRONEM program, Grant No APQ-1452-2.01/10) and by Grants of the National Council of Science and Technology (CNPq No 472533/2013-4 and 474847/2013-6).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Cajueiro, D.B.B., Parente, D.C., Leite, F.C.B. et al. Glutamine: a major player in nitrogen catabolite repression in the yeast Dekkera bruxellensis . Antonie van Leeuwenhoek 110, 1157–1168 (2017). https://doi.org/10.1007/s10482-017-0888-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-017-0888-5