Abstract
Larval frass in herbivorous lepidopterans is mainly composed of plant-derived material and microbes from the gut. Despite the fact that frass from conspecific larvae repels female moths in Spodoptera littoralis from oviposition, the role of frass volatiles on larval foraging behavior is largely unknown. Here, we show that larvae of S. littoralis walk upwind to larval frass volatiles in a wind tunnel assay. We identified the frass volatile guaiacol (2-methoxyphenol) as key ligand for the S. littoralis odorant receptor (OR) SlitOr59 which we expressed heterologously. We isolated guaiacol-producing bacteria identified as Serratia marcescens from frass of larvae that were fed on cotton, and Enterobacter cloacae, E. ludwigii and Klebsiella sp. from frass derived from cabbage-fed larvae. In addition to guaiacol, we also identified volatiles acetoin, 3-methyl-1-butanol and dimethyl disulfide, in large proportions in headspace collections from the bacteria. A Y-tube olfactometer assay showed that fourth instar S. littoralis larvae are attracted to guaiacol. Moreover, cotton leaves treated with the insecticide Spinosad and guaiacol were highly attractive to the larvae. Our results provide a basis for management of the pest by directly targeting larvae, based on an attract-and-kill strategy. Further studies are needed to test the application of guaiacol for semiochemical-based pest management of Spodoptera pest species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Key message
-
We identified an odorant receptor in Spodoptera littoralis (Or59) that responds to guaiacol.
-
Guaiacol-producing bacteria were isolated and identified from larval frass.
-
Larvae are attracted to frass volatiles as well as guaiacol in combination with insecticide.
-
SlitOr59 receptor homology with S. frugiperda SfruOr59 suggests functional similarity.
-
Our results provide a basis for a semiochemical-based pest management strategy.
Introduction
Organic compounds are predominant cues for caterpillars to organize their foraging tactics to locate food resources such as plants (Carlsson et al. 1999; Becher and Guerin 2009). In lepidopterans, as holometabolous insects, behavioral responses to a specific olfactory stimulus can differ strongly between larvae and adults because of their different physiological and ecological requirements. In many lepidopteran species, adults are attracted to floral volatiles for feeding, while larvae mainly are known to respond to leaf volatiles of their host plants. Moreover, differences in larval and adult behavior to host plant volatiles have been shown in noctuid moths, with females being repelled by volatile organic compounds (VOCs) from herbivore-damaged plants (De Moraes et al. 2001; Signoretti et al. 2012), while larvae are attracted to both damaged and undamaged plant volatiles for feeding (Carroll et al. 2006; von Mérey et al. 2013).
The cotton leafworm Spodoptera littoralis (Lepidoptera: Noctuidae) is an important defoliator of many cultivated crops (Salama et al. 1971), and larval feeding results in significant yield reduction (Russell et al. 1993). Defoliation as well as other factors such as predation or unsuitable abiotic conditions leads to larval migration between plants (Martel et al. 2011; von Mérey et al. 2013). To relocate to suitable host plants when crawling on the ground, larvae use reliable olfactory signals such as plant volatiles induced by conspecific feeding (von Mérey et al. 2013). It is unclear whether larvae also use other larval-derived cues to identify suitable host plants. Volatiles emitted from conspecific larval frass that typically is found on the ground of infested plants might help migrating larvae to identify suitable host plants. It was shown that volatiles emitted from frass are perceived by S. littoralis female moths, deterring them from oviposition (Klein et al. 1990), and considering the overlap in larval and adult olfactory systems (Poivet et al. 2013; Walker et al. 2016), it is plausible that frass volatiles might also influence larval behavior. Behavioral responses to insect frass have been shown in other insect species (Dillon et al. 2000; Axelsson et al. 2017; Molnár et al. 2017; Zhang et al. 2019).
Larval frass to a large extent consists of digested plant material and associated microbes which accordingly determine the frass odor. Numerous studies have demonstrated that insects sense microbial-produced VOCs (Dillon et al. 2000; Lauzon et al. 2000; Azeem et al. 2013; Davis and Landolt 2013), and microbes are shown to mediate behavioral responses (Kramer and Abraham 2012; Davis et al. 2013; Becher et al. 2018; Ljunggren et al. 2019). Among the microbes known to induce odor-mediated behavior in insects are those associated with plants (Azeem et al. 2013) and animals (Verhulst et al. 2011) including microbes that are primarily present in the insect gut (Lauzon et al. 2000; Dillon et al. 2002).
An example is the desert locust, Schistocerca gregaria, where the behaviorally active guaiacol (2-methoxyphenol) is produced by the gut bacterium Pantoea agglomerans and emitted from frass (Dillon et al. 2000, 2002). Pantoea agglomerans belongs to the Enterobacteriaceae, and several bacterial species within this family produce aromatic compounds that mediate behavior in insects (Epsky et al. 1998; Lauzon et al. 1998; Axelsson et al. 2017). The larval gut of S. littoralis houses a diverse microbial community that includes various genera within the family Enterobacteriaceae (Tang et al. 2012). We postulate that microbes present in the larval frass of S. littoralis are able to produce volatiles that influence larval behavior and might help them to identify suitable host plants.
Olfaction is one of the main components of chemosensation driving insect behavior, and odorant receptors (ORs) and ionotropic receptors (IRs) are the primary olfactory receptors that detect volatile chemicals (Hansson and Stensmyr 2011). In previous work at our research unit, several ORs of S. littoralis were expressed in a heterologous system to identify OR ligands (de Fouchier et al. 2017). This was achieved following molecular techniques in vivo using Drosophila melanogaster (Δhalo) that lacks its native receptor (Or22a/b within ab3 basiconic sensilla), but retains the expression of the co-receptor (Orco)(Dobritsa et al. 2003). The ‘desired’ OR gene is then expressed within this ‘empty neuron’ following the transgenic techniques (Dobritsa et al. 2003). The process of functionally characterizing the receptor is known as deorphanization (Vosshall et al. 1999; Hallem et al. 2004; Hallem and Carlson 2006). Out of 17 ORs that were functionally characterized in S. littoralis, eight ORs are expressed in both larvae and adults. Yet, as many as 14 out of 22 larval ORs in S. littoralis (Poivet et al. 2013) have not been functionally characterized. Odors that are ecologically relevant are the main candidates to test for behavioral and sensory-physiological activity. Previous panels of test compounds included mainly plant-derived chemicals and pheromones (Rharrabe et al. 2014; de Fouchier et al. 2018). Microbial compounds were previously less prioritized candidates for deorphanization of additional S. littoralis larval ORs.
Here, our objective was to test the following hypotheses: (1) Larval frass of S. littoralis contains microbes that emit VOCs known as fecal volatiles, (2) those microbial volatiles are detected by larval ORs, and (3) these volatiles mediate larval attraction to suitable host plants.
We isolated and identified microbes from frass of S. littoralis larvae fed on three different host plants, and collected their volatiles. Through electrophysiological recordings, we then identified a volatile compound as the key ligand of S. littoralis OR59 (hereafter referred as SlitOr59). Moreover, we determined the compound as a larval attractant in a behavioral assay. Considering the avoidance of frass volatiles by Spodoptera females (Klein et al. 1990), our findings might facilitate development of a ‘push–pull-and-kill’ strategy that repels female moths from oviposition and attracts larvae to a fatal bait.
Materials and methods
Insects and plants
Spodoptera littoralis larvae and adults used in the study were derived from a laboratory colony reared in a climatic chamber at 24 ± 2 °C and 65 ± 5% relative humidity (RH), under a 16:8 h light and dark photoperiod. The eggs of S. littoralis obtained from the laboratory rearing were randomly divided into batches, and hatched larvae were reared on potato-based artificial diet (Hinks and Byers 1976) or one of three different plant species (described below) that have been studied previously as S. littoralis host plants (Thöming et al. 2013). The larval development on artificial diet lasted for 18–21 days consisting of six instars before larvae underwent pupation lasting for 8–10 days. For the behavioral and molecular assays, 10–12-day-old larvae (fourth instar) reared on artificial diet were used. Pupae were separated by sex, and upon adult emergence, the moths were fed on 5% sucrose solution. Adults that were 2–3 days old were mated, and eggs were subsequently used for rearing (Hinks and Byers 1976; Saveer et al. 2012). Larvae were fed on individual host plants (young and mature detached leaves) to collect plant-specific larval frass. The plants used in our studies were cotton (Gossypium hirsutum v. Delta Pineland 90, Malvaceae), cabbage (Brassica oleracea subsp. capitata v. Stonehead, Brassicaceae) and maize (Zea mays v. Tasty Sweet F1, Poaceae). Plants were grown in 1.5 L pots with commercial soil (Hasselfors Garden, Sweden) in climatized growth chambers (Biotron facility at the Swedish University of Agricultural Sciences, Alnarp) at 22 ± 2 °C and 70 ± 5% (RH) under natural daylight supplemented with artificial illumination from sodium lamps.
Isolation and identification of bacterial colonies
Using a sterilized hairbrush, fresh frass was collected from 10–12-day-old larvae that had fed overnight on leaves of individual host plants, such as cotton, cabbage or maize. Approximately, 1 g (n = 3) of frass was collected and added in 1 mL of sterile water, of which 100 μL was further diluted in 900 μL of sterile water. The diluted frass suspension was subsequently used for culturing onto modified nutrient agar plates. The composition of the medium was as follows: glucose (10 g l−1), peptone (5 g l−1), yeast extract (5 g l−1), NaCl (3 g l−1), vanillic acid (1.5 mmol l−1) and agar (15 g l−1) (Dillon et al. 2002). The plates were inoculated with 50 μL of diluted suspension and incubated at 28 °C for 16 h. Three morphologically distinct colonies per host plant (three colonies each for cotton, cabbage and maize) were picked subsequently for bacterial identification and volatile collection.
Bacterial DNA was isolated using the Quick-DNA Fungal/Bacterial Microprep Kit according to manufacturer’s recommendations (Zymo Research, USA). Yield and integrity of the DNA were assessed using a NanoDrop Micro Photometer (NanoDrop Technologies, UK), and agarose gel electrophoresis, respectively. The 16 s rRNA region of all bacterial isolates was PCR amplified individually with the universal primer pairs, 27F (5′- AGAGTTTGATCMTGGCTCAG-3′) (Lane 1991) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′) (Morales and Holben 2009). PCRs were carried out using 10 ng of DNA with the following temperature parameters: initial denaturation step at 94 °C for 3 min, followed by 35 cycles at 94 °C for 45 s, 50 °C for 30 s and 72 °C for 30 s, followed by a final extension step of 72 °C for 5 min. The PCR products were purified using the Qiagen QIAquick PCR Purification Kit (Qiagen, UK). We performed Sanger sequencing for species identification at the GATC biotech AG sequencing facility (Germany) using 27F and 907R primers. DNA star software was used (DNASTAR, USA) to manually analyze and edit nucleotide sequences obtained from the sequencing platform. Resulting sequences with 16 s region were searched for matching hits against the National Center for Biotechnology Information (NCBI) GenBank non-redundant nucleotide database (BLASTn; (Altschul et al. 1997)). Search hits to sequences from records in the database were evaluated for coverage and identity, and the best-matched NCBI accession was recorded. All sequence data are provided in the supplementary material.
RNA extraction from S. littoralis and PCR assays
For RNA extraction of chemosensory tissues, approximately 600 larval heads (200 per replicate) of 10–12-day-old larvae were sampled. In adults, 50 antennal pairs per sex were dissected from 3–4-day-old unmated moths for PCR assays. The TRIzol™ (Thermo Fisher Scientific, Waltham, MA, USA) extraction method was followed to isolate total RNA from the target tissues. Following the manufacturer protocol and the procedure described in Gonzalez et al. (2016), total RNA was precipitated at -20 °C overnight and later washed with 99.9% ethanol. RNA purification was done using DNAse enzyme (TURBO DNA-free™ Kit, Ambion, USA), RDD buffer and the RNeasy MinElute Clean up Kit (Qiagen Kit). The final volume of 50 μL total RNA was eluted using RNase-free water and stored at -20 °C. The purity of RNA was estimated using a NanoDrop spectrophotometer (Thermo Fischer Scientific). For PCR assays and cloning the receptor, cDNA was synthesized using 1 μg total RNA from adults or larvae, respectively.
Volatile collection and chemical analysis
The diluted frass suspension (three replications each per host plant) and bacterial isolates (three isolates for each host plant) were grown in 50 ml vanillic acid medium (Dillon et al. 2002) for 16 h in a shaking incubator at 28 °C at 200 rotations per minute (rpm). The suspension was then poured into a sterile 500-mL wash bottle for headspace sampling. In this system, approximately 0.1 L / min of charcoal-filtered air was pushed to bubble the bacterial suspension through the inlet immersed into the suspension, and the headspace was let to pass through an adsorbent air filter (Super Q, 80/100 mesh; Alltech, Deerfield, IL, USA) at the outlet for 3 h. The adsorbed headspace compounds were eluted with 0.5 ml hexane and used for GC–MS analysis (6890 GC and 5975 MS; Agilent Technologies, USA). Two μL of the sample was injected into a HP-5MS column programmed for 37 min (30 m × 0.25 mm × 0.25 μm film thickness; J&W Scientific, Folsom, CA, USA) with following temperature specifications: initial temperature of 50 °C (2 min) and 8 °C/min to 275 °C (10 min). Compounds were identified based on their Kováts retention indices and mass spectrum using the NIST reference library (Agilent) and standard reference compounds.
Cloning and transgenic expression of SlitOr59
We chose the S. littoralis OR, SlitOr59 (Walker et al. 2019) for cloning and heterologous expression, suspecting that the receptor would be sensitive to aromatic microbial volatiles. One of the principle reasons was that the headspace analysis of medium inoculated with frass obtained from cotton-fed larvae contained several aromatic compounds. Some of these compounds are known to be main ligands of ORs in S. littoralis that have already been deorphanized (de Fouchier et al. 2017). We referred to a phylogenetic tree built on functional characterization of lepidopteran ORs by de Fouchier et al. (2017) as a guide that led us to Bombyx mori Or45 tuned to an aromatic compound, 2-phenylethanol (Tanaka et al. 2009). While, in the same branch, also B. mori OR47 responds to an aromatic compound, surprisingly, no S. littoralis ORs had been characterized from this basal clade. SlitOr59 from S. littoralis clusters with Or45 and Or47 from B. mori among others (Walker et al. 2019), and we therefore hypothesized that SlitOr59 also is physiologically tuned to microbial aromatic compounds. Transcriptomic analysis suggested SlitOr59 expression in larval antennal tissues (Revadi et a., unpublished). Moreover, the receptor is conserved across several Spodoptera species (Gouin et al. 2017; Guo et al. 2020a), and we chose to compare the sequence similarity of SlitOr59 from S. littoralis to the congeneric S. frugiperda OR SfruOr59 (Gouin et al. 2017), by aligning the protein sequences using Clustal Omega (Sievers et al. 2011).
The sequence SlitOr59 was confirmed through PCR amplification followed by Sanger sequencing. The gel-purified PCR product was cloned into the plasmid PCR8/GW/TOPO using the TOPO cloning kit (Thermo Fisher Scientific, USA), and SlitOr59 was inserted into the plasmid. The plasmid was sequenced using GW1 and GW2 primers to confirm desired orientation of the transgene. The sequence was further subcloned into pUASg.attB plasmid (Thermo Fisher, Scientific, USA). The UAS-SlitOr59 lines were generated by BestGene using the PhiC31-mediated integration approach (Chino Hills, CA, USA). Desired D. melanogaster fly lines carrying the SlitOr59 transgene, integrated on the 3rd chromosome: M{3xP3-RFP.attP}ZH-86Fb (with M{vas-int.Dm}ZH-2A) (Bloomington Drosophila Stock Number: 24749), were generated following standard procedure (Gonzalez et al. 2016).
Electrophysiological recordings
Single sensillum recording (SSR)
Electrophysiological recordings were performed on the SlitOr59 receptor expressed in ab3 sensilla of transgenic D. melanogaster flies (n = 6). Flies were prepared for electrophysiological recordings following established procedures (de Fouchier et al. 2017). Briefly, in the recording setup, an immobilized fly was placed approximately 1 cm away from an odor-delivery glass tube connected to a charcoal-filtered humidified airstream (1 L/min). A tungsten reference electrode was manually inserted into the fly eye, while a recording electrode was inserted into the base of the ab3 sensillum using a motor-controlled piezo-micro-manipulator (Märzhauser DC-3 K, Wetzlar, Germany). The signal from the sensillum was amplified (INR-02A and AC/DC UN-06, respectively: Syntech, Kirchzarten, Germany) and transferred to a computer through the IDAC4 interface (Syntech, Kirchzarten, Germany) for visualization. Autospike software (version 3.4, Syntech) was used to analyze the recordings. Odorant compounds diluted in paraffin oil (Merck) to a concentration of 10 μg/μL were used as stock solutions. For recording purposes, odorants were further diluted in paraffin oil to 1:10 and 1: 100 proportions. Disposable Pasteur pipettes (VWR International, Stockholm, Sweden) containing a 1.5 × 0.5 cm filter paper strip (Whitemann) were then loaded with 10 μL of individual diluted odorants to deliver a defined stimulus onto the fly antenna, from which responses were recorded for 0.5 s post-stimulation.
Electroantennography (EAG)
EAG recordings were performed on S. littoralis male and female antennae to determine the responses to guaiacol (n = 6 per sex). Five doses of guaiacol ranging from 10 ng to 100 μg were applied on filter paper (in 10 μL paraffin oil). For recording, the excised moth antennal base was placed at the tip of a glass electrode (filled with Beadle–Ephrussi Ringer solution) connected to a 10 × preamplifier probe that was linked to an IDAC-2 box (Syntech, Kirchzarten, Germany), while the distal end of the antenna was connected to another glass electrode for grounding. The odor delivery system for EAG recording was similar to that of SSR. The output signal from the amplifier was fed into a computer for visualization using the Autospike program (version: 1.2.5, Syntech).
Larval behavioral assays
Wind tunnel larval migration assay
We developed a larval migration assay to test if fecal pellets support host finding in foraging larvae. To mimic natural context, two cotton plants (5–6 weeks old) were kept perpendicularly to wind direction in a wind tunnel (180 X 90 X 60 cm; 30 cm s−1 wind speed) (Supplementary Fig. 1). A Plexiglas plate (24 cm wide X 100 cm long) with a perpendicular cut (4 X 2 cm) in the middle at both longitudinal sides was placed horizontally between the two cotton plants. The plants were slid through the cut into the Plexiglas, and the distance between two plant stems was 16 cm. The height of the Plexiglas plate was adjusted such that the larvae were exposed to the plume of leaf volatiles. Treatments included 0.8 g of larval frass (roughly equivalent to frass from 10 fourth instar larvae feeding on cotton leaves for 24 h) spread (round zones of 8 cm diameter) on the Plexiglas plate around the plant stem, and the same amount of charcoal grains of similar shape as the pellets spread around the control-cotton stem. Larvae that were 10–12 days old were starved overnight prior to the experiments. Larvae were tested individually for 10 min, and their behavior was recorded as ‘treatment’ or ‘control’ responses, or as ‘no choice’ if larvae did not walk upwind, or failed to reach either of the treatment zones (8 cm diameter) in 10 min. In total, 34 larvae were tested, and the treatments were interchanged every fifth larvae to account for positional effects; the Plexiglas surface was cleaned using 70% ethanol.
Spodoptera littoralis fourth instar larval upwind walk to a cotton plant offered with fecal pellets versus a cotton plant with charcoal pellets in a wind tunnel. The data show the responses of larvae to the two treatments and responses of larvae that did not make a choice (mean number ± standard deviation). Asterisks indicate statistically significant differences following a Chi-square test (df = 1; Chi-square = 7.05; p < 0.05; n = 34; no choice = 5)
Y-Olfactometer assay
Two-choice behavioral assays were performed on larvae reared on artificial diet. The size of the Y-olfactometer was: arm length = 14 cm; stem = 12.5 cm; inner diameter = 2.2 cm (developed at Humiglas, Södra Sandby, Sweden). Experiments were performed at 23 ± 2 °C under homogeneous fluorescent light. In order to provide a context to the odorants being tested, two cotton leaf disks of 1 cm diameter were placed on 1 cm2 wet filter papers (Grade 1002, Munktell Filter AB, Munktell) in both arms of the olfactometer. A charcoal-filtered airstream (approx. 0.1 L/min) was humidified using wash bottles containing distilled water and then equally split into the two arms of the olfactometer. Treatments included guaiacol diluted in paraffin oil (100 ng in 10 μL) on the filter paper (1 cm2). The same volume of pure paraffin oil was used as control. To determine the combined effect of guaiacol and insecticide on larval attraction, cotton leaf disks on each side were treated with Spinosad (Laser; Dow Agro Sciences), and the treatments included guaiacol (100 ng in 10 μL) and paraffin oil on the filter paper. Larvae (10–12 days old; starved) were tested individually for 10 min, and depending upon the choice, larval response was recorded as ‘treatment’ or ‘control.’ If larvae did not enter either of the arms of the olfactometer in 10 min, the behavioral response was counted as ‘no-choice’ and excluded from the analysis. The olfactometer arms were flipped every fifth larvae to account for positional effects, and 10–15 larvae were tested per day. After testing 10–15 larvae (each larva as replicate), the olfactometer was washed with water and 70% ethanol and oven-heated at 350 °C overnight.
Statistical analysis
Statistical analyses were performed in R (v. 3.4.3) (R Development Core Team 2017). To account for potential solvent effects in SSR and EAG recordings, we subtracted solvent responses recorded before and after odorant stimulation. Differences in the electrophysiological (SSR) responses (number of spikes) from the empty neuron flies to a panel of compounds (compounds tested at 10 μg dose: o-cresol, guaiacol, 4-methylguaiacol and 2-ethylphenol, and the remaining 24 compounds tested at 100 μg dose) were statistically analyzed by analysis of variance (One-way ANOVA). Tukey’s honestly significant difference (HSD) multiple comparisons were calculated post hoc to test significant differences between treatments (responses to each compound) with confidence interval of 95%. Responses to o-cresol and guaiacol (1 μg and 100 ng) were analyzed by t-test. The intensity of EAG responses in both sexes to five dosages of guaiacol was statistically analyzed by analysis of covariance (ANCOVA). For larval behavioral assays, Pearson's Chi-squared tests were performed to test if the proportion of larvae choosing each side differed from 50:50 at α level = 0.05.
Results
Wind tunnel larval migration assay
In the larval migration assay testing the attraction toward frass in a wind tunnel, a significant higher number of larvae walked upwind toward the cotton plant with fecal pellets compared to the cotton plant with charcoal pellets (df = 1; Chi-square = 6.25; P < 0.05, Fig. 1; Supplementary Fig. 1). Larvae that made a choice, walked perpendicular or diagonally to wind direction before turning into upwind walk. Among larvae that chose the cotton plant with frass, several stayed around the plant until the end of the test. The results confirm our hypothesis that frass helps the larvae to find suitable host plants.
Bacterial identification and GC–MS analyses of microbial volatiles
We extracted and cultured bacteria from frass of larvae fed on three different host plant species. Through Sanger sequencing, we identified four bacterial species representing three genera from nine isolated colonies (Supplementary Table 1). All three bacterial isolates from frass that derived from cotton-fed larvae were identified as Serratia marcescens; from cabbage, we identified Enterobacter cloacae, E. ludwigii and Klebsiella sp.; and from maize, all three isolates were identified as S. grimesii.
We then analyzed and found that all the strains of bacteria produce a variety of different volatile compounds. From the GC–MS analyses, we found differences in the volatile profiles of the medium inoculated with larval frass that derived from host plants or isolated bacteria (Fig. 2; Supplementary Table 2). Medium incubated with frass from different host plants showed high variability in the composition of headspace volatiles. 3-Hydroxy-2-butanone, 3-methyl-1-butanol, 2-methyl-1-butanol and dimethyl disulfide (DMDS) were identified from all incubated frass samples derived from different host plant species. Interestingly, in the headspace derived from medium incubated with frass, guaiacol was identified only in cotton and cabbage samples, but not in samples from maize frass. We also found typical herbivore-induced plant volatiles in the headspace analysis (α-pinene and indole). Moreover, the composition of headspace samples from bacteria that were isolated from cotton- (S. marcescens), cabbage- (E. cloacae, E. ludwigii and Klebsiella sp.) or maize-derived frass (S. grimesii) was partially overlapping with that of medium incubated with frass from larvae fed on the respective plants. Similar to volatiles from incubated frass, isolated bacteria also produced 3-hydroxy-2-butanone, 3-methyl-1-butanol, 2-methyl-1-butanol and DMDS in high proportions. Interestingly, bacteria from larval frass that derived from cotton and cabbage produced high proportions of guaiacol, while bacterial isolates from maize-derived larval frass did not produce guaiacol, but 3-methyl-1-butanol and DMDS in high proportions. (Fig. 2; Supplementary Table 2).
Heatmap of mean relative proportions of compounds identified from the headspace analyses (n = 3) of frass or frass-derived bacteria incubated in growth medium. Frass was sampled from larvae fed on three different host plant species. (K.I. = Kováts retention indices in GC–MS (HP-5 column); S.ma_F31C5 = Serratia marcescens strain F31C5; S.ma_F31C6 = S. marcescens strain F31C6; S.ma_F31C7 = S. marcescens strain F31C7; E.cl = Enterobacter cloacae; E.lu = Enterobacter ludwigii; Kl. = Klebsiella sp.; S.gr_1 = S. grimesii strain MC1; S.gr_2 = S. grimesii strain MC2; S.gr_3 = S. grimesii strain MC3)
Electrophysiological recordings
Identification and deorphanization of the SlitOr59
The position of SlitOr59 in the phylogenetic tree reported by Walker et al. (2019) suggested that the receptor may be sensitive to aromatic compounds (de Fouchier et al. 2017). The RT-PCR analysis revealed that the receptor is expressed in larvae (Supplementary Fig. 2). Therefore, SlitOr59 was selected for further studies and deorphanization. The single sensillum recordings on heterologously expressed SlitOr59 showed that the receptor is highly sensitive to o-cresol and guaiacol followed by 4-methylguaiacol and 2-ethylphenol. The responses to o-cresol and guaiacol were not different from each other (100 ng dose: df = 8.74; t = 1.24; p = 0.25; 1 μg dose: df = 6.9; t = -1.39; p = 0.21). At 10 μg dose, SlitOr59 elicited a highest response to o-cresol and guaiacol compared to 4-methylguaiacol and 2-ethylphenol (ANOVA: df = 3; F = 568.6; p < 0.001). The remaining 24 compounds elicited minor electrophysiological responses at highest dose (100 μg) tested (Fig. 3).
Single sensillum responses of the ab3 sensillum in D. melanogaster carrying the odorant receptor SlitOR59 from S. littoralis. Differences in the electrophysiological responses from the empty neuron flies to a panel of compounds were tested using a t-test, or one-way ANOVA followed by Tukey test for multiple comparisons (n = 6). The top panels are the dose-dependent responses to o-cresol and guaiacol (100 ng (df = 8.74, t = 1.24, p = 0.25) and 1 μg (df = 6.9, t = − 1.39, p = 0.21)), followed by o-cresol and guaiacol, 4-methylguaiacol and 2-ethylphenol (10 μg (ANOVA: df = 3, F = 568.6, p < 0.001)) and all other phenolic compounds tested at the highest doses (100 μg (ANOVA: df = 25, F = 259.7, p < 0.001)). The neuronal activity was recorded for 28 compounds in total that mainly comprised of aromatic compounds. Two compounds, o-cresol and guaiacol, showed higher responses compared to the rest of the compounds
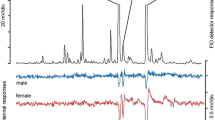
Responses to guaiacol in adult moth antennae
SlitOr59 was confirmed to be expressed in the antennae of adult moths too (Supplementary Fig. 2). We therefore checked electrophysiological responses of adult antennae by EAG recordings (Fig. 4). We found that S. littoralis adults of both sexes respond to guaiacol in a dose-dependent way and that females respond stronger than males (ANCOVA: Concentration df = 4; F = 142.61; p < 0.001; Sex df = 1; F = 17.13; p < 0.001, interaction (Concentration*Sex) df = 4; F = 3.73; p < 0.05; residual df = 50).
Dose-dependent EAG responses (expressed in millivolts) for S. littoralis male and female severed antennae to five doses of guaiacol (n = 6 per sex). (ANCOVA: Concentration df = 4; F = 142.61; p < 0.001; Sex df = 1; F = 17.13; p < 0.001, interaction (Concentration*Sex) df = 4; F = 3.73; p < 0.05; residual df = 50). The amplitudes for the control stimulus (paraffin oil) before and after odorant stimulation were subtracted from the absolute EAG amplitudes. The boxplot represents mean values with the lower and the upper end point of the confidence interval at 95%. Outliers are marked as green (for male) and black (for female) dots
Olfactometer assays
In positive control tests, a significant number of larvae preferred the odor of artificial diet to a blank control of purified air (df = 1; Chi-square = 17.64; P < 0.001) (Fig. 5). Furthermore, a significant number of larvae preferred cotton leaf disks to blank control air (df = 1; Chi-square = 16.95; P < 0.001). In a third control test, no significant difference was found in attraction to cotton leaf disks offered in both sides of the olfactometer (df = 1; Chi-square = 0.36; P = 0.55). With cotton leaf disks as background, larvae highly preferred guaiacol dissolved in paraffin oil over paraffin oil (df = 1; Chi-square = 11.22; P < 0.001) when tested for attraction to 100 ng guaiacol. Corroborating this result, a combination of Spinosad-treated cotton leaf disks and 100 ng guaiacol attracted a significant number of larvae over Spinosad-treated cotton leaf disks by itself (df = 1; Chi-square = 7.05; P < 0.01).
Behavioral responses of S. littoralis larva in Y-tube olfactometer. The data show the percentage of larvae choosing each arm for five different odor pairs: artificial diet vs control blank (df = 1; Chi-square = 17.64; p < 0.001; n = 25; no choice = 2); cotton leaf disk vs clean air (df = 1; Chi-square = 16.95; p < 0.001; n = 48; no choice = 5); cotton leaf disk vs cotton leaf disk (df = 1; Chi-square = 0.36; p = 0.55; n = 25; no choice = 0); cotton leaf disk + guaiacol in paraffin oil vs cotton leaf disk + paraffin oil (df = 1; Chi-square = 11.22; p < 0.001; n = 65; no choice = 3); cotton leaf disk + guaiacol in paraffin oil + Spinosad vs cotton leaf disk + paraffin oil + Spinosad (df = 1; Chi-square = 7.05; p < 0.01; n = 41; no choice = 0). Asterisks indicate statistically significant differences following a Chi-square test. (*** = p < 0.001; ** = p < 0.01)
Discussion
In this study, we demonstrated that S. littoralis larvae are attracted to guaiacol. We postulated that microbes present in the larval frass of S. littoralis are able to produce volatiles that influence larval behavior and might help them to identify suitable host plants. In agreement with our postulation, larvae walked upwind to cotton plants in response to larval frass placed around the plant stem. Frass volatiles from larvae most likely indicate the presence of conspecific larval feeding as a reliable olfactory signal for a suitable food resource. We were able to trace the production of guaiacol to culturable bacteria in frass of larvae fed on cotton and cabbage leaves. Frass-derived compounds such as acetoin, guaiacol and 1-octanol (Klein et al. 1990) were detected in headspace of isolated bacterial colonies and confirmed that larval frass contains microbial communities that emit VOCs, contributing to fecal volatile emissions.
We identified an odorant receptor SlitOr59 in S. littoralis that is physiologically sensitive to guaiacol, a volatile component previously reported from larval frass (Klein et al. 1990). This is the first OR within lepidopterans described for responding to guaiacol. Previously, few insect ORs have been described to respond to guaiacol in D. melanogaster (Hallem and Carlson 2006; Dweck et al. 2015) and S. gregaria (Guo et al. 2020b). In S. littoralis, the guaiacol-sensitive OR also responds to similarly structured compounds, such as o-cresol, 4-methylguaiacol and 2-ethylphenol.
Guaiacol perception in S. littoralis parallels guaiacol perception in other insects, including locusts (Dillon et al. 2000), flies (Drosophila melanogaster) (Hallem and Carlson 2006), mosquitoes (Anopheles gambiae) (Hallem et al. 2004), weevils (Hylobius abietis) (Axelsson et al. 2017) and beetles (Carpophilus humeralis) (Zilkowski et al. 1999), although the receptors across insect orders are generally phylogenetically unrelated (Hansson and Stensmyr 2011). Accordingly, sensitivity to guaiacol across insect orders suggests convergent evolution. Furthermore, within Lepidoptera phylogeny of ORs, SlitOr59 falls into a clade of ORs that is generally more conserved than more divergent ORs such as those that mediate responses to pheromone (de Fouchier et al. 2017; Walker et al. 2019). The identification of guaiacol as key ligand for SlitOR59 proves the validity of predicting OR ligands based on OR phylogeny and molecular structure of respective odor compounds. Both physiological and behavioral responses highlight the importance of microbial-derived volatiles such as guaiacol across diverse insect taxa.
Spodoptera littoralis is naturally exposed to different microbial communities via feeding on leaves and other contacts with the environment shaping the microbial community in the gut (Voirol et al. 2018). The microbial community in the S. littoralis larval gut is known to be dynamic (Shao et al. 2014; Chen et al. 2016). In caterpillars, gut microbes generally show low abundance and are mainly transient, leaf-derived passengers excreted with frass (Hammer et al. 2017). We were able to isolate and identify numerically dominant microbes from frass derived from cotton, cabbage and maize representing five species of three bacterial genera.
In situ functions of gut microbes in lepidopterans and other insects are increasingly studied, especially functions related to plant feeding, digestion and nutrient acquisition (Hammer et al. 2017; Ankrah and Douglas 2018; Voirol et al. 2018). While on the one hand caterpillars are discussed to show high independence from gut microbes (Hammer et al. 2017), other studies suggest important microbial functions related to immunity (Shao et al. 2017), digestion and detoxification of plant compounds (Xia et al. 2017). Interestingly, in Plutella xylostella, functional identification of larval gut microbes showed Enterobacter sp. (E. cloacae and E. asburiae) as dominant bacteria, producing a series of enzymes that are vital for digestion, amino acid synthesis and detoxification of plant defense compounds (Xia et al. 2017). Our finding of E. cloacae and E. ludwigii in the frass of larvae fed on cabbage suggests that these microbes might have similar biological functions in S. littoralis larvae. Interestingly, in S. frugiperda, antibiotic-treated larvae or their oral regurgitant induced higher anti-herbivore defense protein activities (polyphenol oxidase expressed in milligrams per milliliter) in tomato plants compared to untreated larvae (Acevedo et al. 2017). The oral secretions of untreated larvae contained herbivore-induced defense suppressing bacteria identified as P. ananatis and Enterobacteriaceae-1 (genus Serratia/Rahnella) that are involved in regulating plant defense (Acevedo et al. 2017). In the future, similar studies in S. littoralis using antibiotics are needed to understand the role of guaiacol-producing bacteria.
In maize-fed larval frass, we identified S. grimesii, a bacterium known to be associated with maize roots (Prischmann et al. 2008). Our volatile analyses showed that S. grimesii from maize-derived frass produced large amounts of DMDS, a volatile also known from frass of other lepidopteran species (Auger et al. 1989; Agelopoulos et al. 1995; Reddy et al. 2002). In cabbage plants, disrupted tissues and roots infested with Delia radicum larvae emit high amounts of DMDS (Chin and Lindsay 1993; Ferry et al. 2007) and Serratia species inside the larvae evidently account for the synthesis of the sulfur volatile (Lukwinski et al. 2006; van Dam et al. 2012). Noteworthy, DMDS is another example of volatiles produced by plants and microorganisms. It is intriguing to find distinct volatile compounds produced by plant tissue and its associated microbes. Several studies on plant-microbial interactions illustrate that plants in nature interact with multitude of microorganisms on leaves and root surfaces, of which several of them promote plant health (Hacquard et al. 2017). The assembly of microorganisms in the plant phyllosphere is not random but influenced by plant species and environmental factors (Vorholt 2012; Thapa and Prasanna 2018; Darlison et al. 2019). The overlapping VOCs in plants and microbes exemplify that microorganisms contribute essentially to plant volatile emissions as previously discussed for flowers and fruits (Stökl et al. 2010; Nasopoulou et al. 2014; Becher et al. 2018).
The bacteria that were isolated from feces of larvae fed on cotton or cabbage were able to produce guaiacol. Guaiacol and other bacterial volatiles detected in our study have been previously identified as insect frass components in different insect species and function as oviposition deterrents in moths (benzyl alcohol, guaiacol, acetophenone) (Klein et al. 1990; Molnár et al. 2017), antifeedant effectors in weevils (guaiacol, 4-methylanisole, styrene, 2-phenylethanol) (Borg-Karlson et al. 2006; Azeem et al. 2013; Axelsson et al. 2017), kairomones for braconid parasitoid wasps (guaiacol) (Ramachandran et al. 1991) and as behavioral inhibitor in locusts (guaiacol) (Guo et al. 2020b).
Klein et. al. (1990) found as many as 78 compounds in larval frass of S. littoralis. Some of the compounds overlap with that of herbivore induced volatiles in cotton plants (Binyameen et al. 2014). The difference in composition between plant and frass odor blends is characterized by microbial compounds such as guaiacol, acetoin and 1-octanol (Klein et al. 1990; Binyameen et al. 2014). In our headspace analysis of frass suspensions, we detected microbial volatiles and some of the herbivore-induced plant volatiles, such as isoamyl acetate, alpha-pinene and (E)-β-ocimene (Binyameen et al. 2014). In conclusion, larval frass emits a mixture of microbial volatiles and a subset of induced plant volatiles that mediate attraction in larvae. We propose that microbes and their volatile metabolites in S. littoralis larval frass help larvae to identify conspecific feeding sites.
Spodoptera littoralis larval-expressed ORs such as SlitOR24, SlitOR25 and SlitOR36 are shown to be physiologically sensitive to frass volatiles such as, benzaldehyde, benzyl alcohol, acetophenone and 1-indanone (Klein et al. 1990; de Fouchier et al. 2017). Benzaldehyde, benzyl alcohol and acetophenone were known as frass-derived compounds behaviorally active in larvae (de Fouchier et al. 2018). We found guaiacol as a main compound in the media inoculated with larval frass, behaviorally active in larvae and eliciting strong physiological response in SlitOr59. Although guaiacol-containing frass is known as an oviposition deterrent with respect to S. littoralis female moth behavior (Klein et al. 1990), physiological responses from the moth antennae to guaiacol have not been previously established. High sensitivity in the female antennae suggests that guaiacol detection might influence egg-laying decisions. In another lepidopteran moth, Cydalima perspectalis, guaiacol elicited higher physiological responses in females compared to male moth EAG antennal recordings and serves as oviposition deterrent (Molnár et al. 2017).
The olfactory system in larvae of S. littoralis appears to be adapted to not only encode plant volatiles but also microbial VOCs in the environment. In an ecological context, interactions between larvae and several bacterial species producing guaiacol appear complex. Further studies are needed to determine in situ functions of these microbes in the gut. Indeed, the interactions appear beneficial for larvae, as they may help larvae find suitable feeding sites, with detrimental consequences for the plant. Interestingly, for the closely related congeneric species, S. frugiperda, it was shown that frass-derived proteins suppress herbivore-induced defense in maize, thereby increasing larval performance (Ray et al. 2016).
Our data suggest that guaiacol combined with insecticide might be used as an attract-and-kill method for control of S. littoralis larvae. The female antennal responses to guaiacol in our study and the previously described oviposition deterrence to guaiacol containing frass (Klein et al. 1990) provide a basis for sustainable pest control directly targeting the larvae and adults, opening up a new strategy of push–pull and kill. Interestingly, natural enemies such as predators and parasitoids also use frass volatiles to locate their prey (Ramachandran et al. 1991; Reddy et al. 2002). For example, the parasitoid, Microplitis demolitor (Braconidae: Hymenoptera), is attracted to its host, Pseudoplusia includens (Noctuidae) through larval frass volatiles, such as guaiacol and 3-octanone (Ramachandran et al. 1991). The manipulation of the pest with frass-derived semiochemicals might accordingly be compatible with biological control.
The phylogenetic tree of Spodoptera ORs described in Guo et al. (2020a) shows that the S. frugiperda olfactory receptor (SfruOr59) and S. littoralis SlitOr59 are homologs, suggesting a similar response pattern. Moreover, sequence alignment of SlitOr59 and SfruOr59 performed using Clustal Omega (1.2.4) (Sievers et al. 2011) showed 92.91% similarity (Supplementary Fig. 3). Considering the receptor homology, guaiacol with insecticide treatment might not only be of use to control S. littoralis, but even to manage S. frugiperda. Further studies are needed to test whether guaiacol improves the efficiency of pesticide application.
Author contributions
SVR and PGB conceived the idea and wrote the manuscript. SVR, RRV, WWB and PGB designed the experiments. SVR, RRV and WWB collected molecular biology data. SVR, VAG and PGB collected behavioral experiment data. SVR analyzed the data and wrote the first manuscript draft. All authors commented and approved the manuscript.
Availability of data and materials
All supplementary material and data is available.
References
Acevedo FE, Peiffer M, Tan C-W et al (2017) Fall armyworm-associated gut bacteria modulate plant defense responses. Mol Plant Microbe Interact 30:127–137. https://doi.org/10.1094/MPMI-11-16-0240-R
Agelopoulos NG, Dicke M, Posthumus MA (1995) Role of volatile infochemicals emitted by feces of larvae in host-searching behavior of parasitoid Cotasia rubecula (Hymenoptera: Braconidae): a behavioral and chemical study. J Chem Ecol 21:1789–1811. https://doi.org/10.1007/BF02033677
Altschul SF, Madden TL, Schäffer AA et al (1997) Gapped BLAST and PS I-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Ankrah NYD, Douglas AE (2018) Nutrient factories: metabolic function of beneficial microorganisms associated with insects. Environ Microbiol 20:2002–2011. https://doi.org/10.1111/1462-2920.14097
Auger J, Lecomte C, Paris J, Thibout E (1989) Identification of leek-moth and diamondback-moth frass volatiles that stimulate parasitoid, Diadromus pulchellus. J Chem Ecol 15:1391–1398. https://doi.org/10.1007/BF01014838
Axelsson K, Konstanzer V, Rajarao GK et al (2017) Antifeedants produced by bacteria associated with the gut of the pine weevil Hylobius abietis. Microb Ecol 74:177–184. https://doi.org/10.1007/s00248-016-0915-5
Azeem M, Rajarao GK, Nordenhem H et al (2013) Penicillium expansum volatiles reduce pine weevil attraction to host plants. J Chem Ecol 39:120–128. https://doi.org/10.1007/s10886-012-0232-5
Becher PG, Guerin PM (2009) Oriented responses of grapevine moth larvae Lobesia botrana to volatiles from host plants and an artificial diet on a locomotion compensator. J Insect Physiol 55:384–393. https://doi.org/10.1016/j.jinsphys.2009.01.006
Becher PG, Hagman A, Verschut V et al (2018) Chemical signaling and insect attraction is a conserved trait in yeasts. Ecol Evol 8:2962–2974. https://doi.org/10.1002/ece3.3905
Binyameen M, Anderson P, Ignell R et al (2014) Identification of plant semiochemicals and characterization of new olfactory sensory neuron types in a polyphagous pest moth, Spodoptera littoralis. Chem Senses 39:719–733. https://doi.org/10.1093/chemse/bju046
Borg-Karlson AK, Nordlander G, Mudalige A et al (2006) Antifeedants in the feces of the pine weevil Hylobius abietis: Identification and biological activity. J Chem Ecol 32:943–957. https://doi.org/10.1007/s10886-006-9050-y
Carlsson MA, Anderson P, Hartlieb E, Hansson BS (1999) Experience-dependent modification of orientational response to olfactory cues in larvae of Spodoptera littoralis. J Chem Ecol 25:2445–2454. https://doi.org/10.1023/A%3A1020865922827
Carroll MJ, Schmelz EA, Meagher RL, Teal PEA (2006) Attraction of Spodoptera frugiperda larvae to volatiles from herbivore-damaged maize seedlings. J Chem Ecol 32:1911–1924. https://doi.org/10.1007/s10886-006-9117-9
Chen B, Teh B-S, Sun C et al (2016) Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci Rep 6:29505. https://doi.org/10.1038/srep29505
Chin H-W, Lindsay RC (1993) Volatile sulfur compounds formed in disrupted tissues of different cabbage cultivars. J Food Sci 58:835–839. https://doi.org/10.1111/j.1365-2621.1993.tb09370.x
Darlison J, Mogren L, Rosberg AK et al (2019) Leaf mineral content govern microbial community structure in the phyllosphere of spinach (Spinacia oleracea) and rocket (Diplotaxis tenuifolia). Sci Total Environ 675:501–512. https://doi.org/10.1016/j.scitotenv.2019.04.254
Davis TS, Crippen TL, Hofstetter RW, Tomberlin JK (2013) Microbial volatile emissions as insect semiochemicals. J Chem Ecol 39:840–859. https://doi.org/10.1007/s10886-013-0306-z
Davis TS, Landolt PJ (2013) A survey of insect assemblages responding to volatiles from a ubiquitous fungus in an agricultural landscape. J Chem Ecol 39:860–868. https://doi.org/10.1007/s10886-013-0278-z
de Fouchier A, Sun X, Caballero-Vidal G et al (2018) Behavioral effect of plant volatiles binding to Spodoptera littoralis larval odorant receptors. Front Behav Neurosci 12:264. https://doi.org/10.3389/fnbeh.2018.00264
de Fouchier A, Walker WB, Montagne N et al (2017) Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat Commun 8:15709. https://doi.org/10.1038/ncomms15709
De Moraes CM, Mescher MC, Tumlinson JH (2001) Caterpillar-induced nocturnal plant volatiles repel conspecific females. Nature 410:577–580. https://doi.org/10.1038/35069058
Dillon RJ, Vennard CT, Charnley AK (2002) A note: Gut bacteria produce components of a locust cohesion pheromone. J Appl Microbiol 92:759–763. https://doi.org/10.1046/j.1365-2672.2002.01581.x
Dillon RJ, Vennard CT, Charnley AK (2000) Exploitation of gut bacteria in the locust. Nature 403:851. https://doi.org/10.1016/j.echo.2007.08.009
Dobritsa AA, Der Goes V, Van Naters W, Warr CG et al (2003) Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37:827–841. https://doi.org/10.1016/S0896-6273(03)00094-1
Dweck HKM, Ebrahim SAM, Farhan A et al (2015) Olfactory proxy detection of dietary antioxidants in Drosophila. Curr Biol 25:455–466. https://doi.org/10.1016/j.cub.2014.11.062
Epsky ND, Heath RR, Dueben BD et al (1998) Attraction of 3-methyl-1-butanol and ammonia identified from Enterobacter agglomerans to Anastrepha suspensa. J Chem Ecol 24:1867–1880. https://doi.org/10.1023/A:1022363718193
Ferry A, Dugravot S, Delattre T et al (2007) Identification of a widespread monomolecular odor differentially attractive to several Delia radicum ground-dwelling predators in the field. J Chem Ecol 33:2064–2077. https://doi.org/10.1007/s10886-007-9373-3
Gonzalez F, Witzgall P, Walker WB (2016) Protocol for heterologous expression of insect odourant receptors in Drosophila. Front Ecol Evol 4:1–15. https://doi.org/10.3389/fevo.2016.00024
Gouin A, Bretaudeau A, Nam K et al (2017) Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci Rep 7:1–12. https://doi.org/10.1038/s41598-017-10461-4
Guo JM, Liu XL, Liu SR et al (2020a) Functional characterization of sex pheromone receptors in the fall armyworm (Spodoptera frugiperda). Insects. https://doi.org/10.3390/insects11030193
Guo X, Yu Q, Chen D et al (2020b) 4-Vinylanisole is an aggregation pheromone in locusts. Nature 584:584–588. https://doi.org/10.1038/s41586-020-2610-4
Hacquard S, Spaepen S, Garrido-Oter R, Schulze-Lefert P (2017) Interplay between Innate Immunity and the Plant Microbiota. Annu Rev Phytopathol 55:565–589. https://doi.org/10.1146/annurev-phyto-080516-035623
Hallem EA, Carlson JR (2006) Coding of odors by a receptor repertoire. Cell 125:143–160. https://doi.org/10.1016/j.cell.2006.01.050
Hallem EA, Nicole Fox A, Zwiebel LJ, Carlson JR (2004) Olfaction: mosquito receptor for human-sweat odorant. Nature 427:212–213. https://doi.org/10.1038/427212a
Hammer TJ, Janzen DH, Hallwachs W et al (2017) Caterpillars lack a resident gut microbiome. Proc Natl Acad Sci U S A 114:9641–9646. https://doi.org/10.1073/pnas.1707186114
Hansson BS, Stensmyr MC (2011) Evolution of insect olfaction. Neuron 72:698–711. https://doi.org/10.1016/j.neuron.2011.11.003
Hinks CF, Byers JR (1976) Biosystematics of the genus Euxoa (Lepidoptera: Noctuidae) v. Rearing procedures, and life cycles of 36 species. Can Entomol 108:1345–1357. https://doi.org/10.4039/Ent1081345-12
Klein B, Schildknecht H, Hilker M, Bombosch S (1990) Eiablagehemmende Wirkstoffe aus dem Larvenkot von Spodoptera littoralis (Boisd). Zeitschrift für Naturforsch C 45c:895–901. https://doi.org/10.1080/03235408.2012.755047
Kramer R, Abraham WR (2012) Volatile sesquiterpenes from fungi: What are they good for? Phytochem Rev 11:15–37. https://doi.org/10.1007/s11101-011-9216-2
Lane DJ (1991) 16S/23S rRNA Sequencing. Nucleic acid Tech Bact Syst. https://doi.org/10.1007/s00227-012-2133-0
Lauzon CR, Sjogren RE, Prokopy RJ (2000) Enzymatic capabilities of bacteria associated with apple maggot flies: a postulated role in attraction. J Chem Ecol 26:953–967. https://doi.org/10.1023/A:1005460225664
Lauzon CR, Sjogren RE, Wright SE, Prokopy RJ (1998) Attraction of Rhagoletis pomonella (Diptera: Tephritidae) flies to odor of bacteria: apparent confinement to specialized members of Enterobacteriaceae. Environ Entomol 27:853–857. https://doi.org/10.1093/ee/27.4.853
Ljunggren J, Borrero-Echeverry F, Chakraborty A et al (2019) Yeast volatomes differentially effect larval feeding in an insect herbivore. Appl Environ Microbiol 85:1–16. https://doi.org/10.1128/aem.01761-19
Lukwinski AT, Hill JE, Khachatourians GG et al (2006) Biochemical and taxonomic characterization of bacteria associated with the crucifer root maggot (Delia radicum). Can J Microbiol 52:197–208. https://doi.org/10.1139/w05-123
Martel V, Schlyter F, Ignell R et al (2011) Mosquito feeding affects larval behaviour and development in a moth. PLoS ONE 6:1–5. https://doi.org/10.1371/journal.pone.0025658
Molnár BP, Tóth Z, Kárpáti Z (2017) Synthetic blend of larval frass volatiles repel oviposition in the invasive box tree moth, Cydalima perspectalis. J Pest Sci 90:873–885. https://doi.org/10.1007/s10340-017-0837-0
Morales SE, Holben WE (2009) Empirical testing of 16S rRNA gene PCR primer pairs reveals variance in target specificity and efficacy not suggested by in silico analysis. Appl Environ Microbiol 75:2677–2683. https://doi.org/10.1128/AEM.02166-08
Nasopoulou C, Pohjanen J, Koskimäki JJ et al (2014) Localization of strawberry (Fragaria x ananassa) and Methylobacterium extorquens genes of strawberry flavor biosynthesis in strawberry tissue by in situ hybridization. J Plant Physiol 171:1099–1105. https://doi.org/10.1016/j.jplph.2014.03.018
Poivet E, Gallot A, Montagné N et al (2013) A Comparison of the olfactory gene repertoires of adults and larvae in the noctuid moth Spodoptera littoralis. PLoS ONE. https://doi.org/10.1371/journal.pone.0060263
Prischmann DA, Lehman RM, Christie AA, Dashiell KE (2008) Characterization of bacteria isolated from maize roots: Emphasis on Serratia and infestation with corn rootworms (Chrysomelidae: Diabrotica). Appl Soil Ecol 40:417–431. https://doi.org/10.1016/j.apsoil.2008.06.012
R Development Core Team R (2017) R: A language and environment for statistical computing. R Found. Stat. Comput.
Ramachandran R, Norris DM, Phillips JK, Phillips TW (1991) Volatiles mediating plant-herbivore-natural enemy interactions: Soybean looper frass volatiles 3-octanone and guaiacol as kairomones for the parasitoid Microplitis demolitor. J Agric Food Chem 39:2310–2317. https://doi.org/10.1021/jf00012a044
Ray S, Alves PCMS, Ahmad I et al (2016) Turnabout is fair play: Herbivory-induced plant chitinases excreted in fall armyworm frass suppress herbivore defenses in maize. Plant Physiol 171:694–706. https://doi.org/10.1104/pp.15.01854
Reddy GVP, Holopainen JK, Guerrero A (2002) Olfactory responses of Plutella xylostella natural enemies to host pheromone, larval frass, and green leaf cabbage volatiles. J Chem Ecol 28:131–143. https://doi.org/10.1023/A:1013519003944
Rharrabe K, Jacquin-Joly E, Marion-Poll F (2014) Electrophysiological and behavioral responses of Spodoptera littoralis caterpillars to attractive and repellent plant volatiles. Front Ecol Evol 2:1–9. https://doi.org/10.3389/fevo.2014.00005
Russell DA, Radwan SM, Irving NS et al (1993) Experimental assessment of the impact of defoliation by Spodoptera littoralis on the growth and yield of giza ’75 cotton. Crop Prot 12:303–309. https://doi.org/10.1016/0261-2194(93)90051-J
Salama HS, Dimetry NZ, Salem SA (1971) On the host preference and biology of the cotton leafworm Spodoptera littoralis Bois. Zeitschrift für Angew Entomol 67:261–266. https://doi.org/10.1111/j.1439-0418.1971.tb02122.x
Saveer AM, Kromann SH, Birgersson G et al (2012) Floral to green: mating switches moth olfactory coding and preference. Proc R Soc B Biol Sci 279:2314–2322. https://doi.org/10.1098/rspb.2011.2710
Shao Y, Arias-Cordero E, Guo H et al (2014) In vivo Pyro-SIP assessing active gut microbiota of the cotton leafworm, Spodoptera littoralis. PLoS ONE 9:1–13. https://doi.org/10.1371/journal.pone.0085948
Shao Y, Chen B, Sun C et al (2017) Symbiont-derived antimicrobials contribute to the control of the Lepidopteran gut microbiota. Cell Chem Biol 24:66–75. https://doi.org/10.1016/j.chembiol.2016.11.015
Sievers F, Wilm A, Dineen D et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. https://doi.org/10.1038/msb.2011.75
Signoretti AGC, Peñaflor MFGV, Bento JMS (2012) Fall Armyworm, Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae), female moths respond to herbivore-induced corn volatiles. Neotrop Entomol 41:22–26. https://doi.org/10.1007/s13744-011-0003-y
Stökl J, Strutz A, Dafni A et al (2010) A deceptive pollination system targeting drosophilids through olfactory mimicry of yeast. Curr Biol 20:1846–1852. https://doi.org/10.1016/j.cub.2010.09.033
Tanaka K, Uda Y, Ono Y et al (2009) Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr Biol 19:881–890. https://doi.org/10.1016/j.cub.2009.04.035
Tang X, Freitak D, Vogel H et al (2012) Complexity and variability of gut commensal microbiota in polyphagous lepidopteran larvae. PLoS ONE 7:1–9. https://doi.org/10.1371/journal.pone.0036978
Thapa S, Prasanna R (2018) Prospecting the characteristics and significance of the phyllosphere microbiome. Ann Microbiol 68:229–245. https://doi.org/10.1007/s13213-018-1331-5
Thöming G, Larsson MC, Hansson BS, Anderson P (2013) Comparison of plant preference hierarchies of male and female moths and the impact of larval rearing hosts. Ecology 94:1744–1752. https://doi.org/10.1890/12-0907.1
van Dam NM, Samudrala D, Harren FJM, Cristescu SM (2012) Real-time analysis of sulfur-containing volatiles in Brassica plants infested with root-feeding Delia radicum larvae using proton-transfer reaction mass spectrometry. AoB Plants 2012:1–12. https://doi.org/10.1093/aobpla/pls021
Verhulst NO, Qiu YT, Beijleveld H et al (2011) Composition of human skin microbiota affects attractiveness to malaria mosquitoes. PLoS ONE 6:e28991. https://doi.org/10.1371/journal.pone.0028991
Voirol LRP, Frago E, Kaltenpoth M et al (2018) Bacterial symbionts in lepidoptera: Their diversity, transmission, and impact on the host. Front Microbiol 9:1–14. https://doi.org/10.3389/fmicb.2018.00556
von Mérey GE, Veyrat N, D’Alessandro M, Turlings TCJ (2013) Herbivore-induced maize leaf volatiles affect attraction and feeding behavior of Spodoptera littoralis caterpillars. Front Plant Sci 4:209. https://doi.org/10.3389/fpls.2013.00209
Vorholt JA (2012) Microbial life in the phyllosphere. Nat Rev Microbiol 10:828–840. https://doi.org/10.1038/nrmicro2910
Vosshall LB, Amrein H, Morozov PS et al (1999) A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96:725–736. https://doi.org/10.1016/S0092-8674(00)80582-6
Walker WB, Gonzalez F, Garczynski SF, Witzgall P (2016) The chemosensory receptors of codling moth Cydia pomonella expression in larvae and adults. Sci Rep 6:23518. https://doi.org/10.1038/srep23518
Walker WB, Roy A, Anderson P et al (2019) Transcriptome analysis of gene families involved in chemosensory function in Spodoptera littoralis (Lepidoptera: Noctuidae). BMC Genomics 20:428. https://doi.org/10.1186/s12864-019-5815-x
Xia X, Gurr GM, Vasseur L et al (2017) Metagenomic sequencing of Diamondback moth gut microbiome unveils key holobiont adaptations for herbivory. Front Microbiol 8:1–12. https://doi.org/10.3389/fmicb.2017.00663
Zhang J, Bisch-Knaden S, Fandino RA, Yan S, Obiero GF, Grosse-Wilde E, Hansson BS, Knaden M (2019) The olfactory coreceptor IR8a governs larval feces-mediated competition avoidance in a hawkmoth. Proc Nat Acad Sci 116 (43):21828–21833
Zilkowski BW, Bartelt RJ, Blumberg D et al (1999) Identification of host-related volatiles attractive to pineapple beetle Carpophilus humeralis. J Chem Ecol 25:229–252. https://doi.org/10.1023/A:1020857721010
Acknowledgements
This study was supported by the Linnaeus initiative `Insect Chemical Ecology, Ethology, and Evolution (IC-E3)’ through the Swedish Research Council Formas and SLU, and Formas grants to SR (Grant 2016-01434) and PGB (Grant 2015-1221). PGB was supported by the SLU Centre for Biological Control (CBC). The views expressed herein do not necessarily reflect the official opinion of the donors.
Funding
Open access funding provided by Swedish University of Agricultural Sciences.. This study was supported by the Linnaeus initiative `Insect Chemical Ecology, Ethology, and Evolution (IC-E3)’ through the Swedish Research Council Formas and SLU, and Formas grants to SR (Grant 2016–01434) and PGB (Grant 2015–1221). PGB was supported by the SLU Centre for Biological Control (CBC).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Communicated by N. Desneux .
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10340_2021_1352_MOESM1_ESM.pdf
Supplementary Figure 1: Graphical representation of S. littoralis larval migration assay in the wind tunnel. (PDF 87 KB)
10340_2021_1352_MOESM2_ESM.tif
Supplementary Figure 2: Reverse transcription PCR (RT-PCR) assay for SlitOR59 expression in S. littoralis larvae and adults: The assay was performed using gene-specific primers and cDNA from larval and adult chemosensory tissue: Control, larva, male and female. (TIF 219 KB)
10340_2021_1352_MOESM3_ESM.jpeg
Supplementary Figure 3: Amino acid alignment of SlitOr59 from S. littoralis and SfruOr59 from S. frugiperda using Clustal Omega (Needleman-Wunsch algorithm). In the figure, vertical bar (|) indicate similarity and fully conserved amino acids, (:) indicate highly conserved amino acids with similar properties, and (.) indicate weakly conserved amino acids.
10340_2021_1352_MOESM4_ESM.docx
Supplementary Table 1: Bacterial strains identified from feces of larvae of S. littoralis fed on three different host plants. Morphologically typical isolates from each host plants were sequenced using Sanger sequencing platform. (DOCX 18 KB)
10340_2021_1352_MOESM5_ESM.xlsx
Supplementary Table 2: Mean relative proportion of compounds identified from the headspace analysis (n = 3) of frass or frass-derived bacteria incubated in growth medium. Frass was sampled from larvae fed on three different host plant species. (K.I. = Kováts retention indices in GC-MS (HP-5 column, mobile phase helium); (XLSX 12 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Revadi, S.V., Giannuzzi, V.A., Vetukuri, R.R. et al. Larval response to frass and guaiacol: detection of an attractant produced by bacteria from Spodoptera littoralis frass. J Pest Sci 94, 1105–1118 (2021). https://doi.org/10.1007/s10340-021-01352-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10340-021-01352-9