Abstract
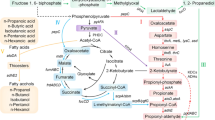
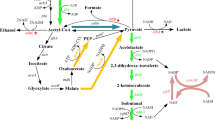
Higher alcohols, longer chain alcohols, contain more than 3 carbon atoms, showed close energy advantages as gasoline, and were considered as the next generation substitution for chemical fuels. Higher alcohol biosynthesis by native microorganisms mainly needs gene expression of heterologous keto acid decarboxylase and alcohol dehydrogenases. In the present study, branched-chain α-keto acid decarboxylase gene from Lactococcus lactis subsp. lactis CICC 6246 (Kivd) and alcohol dehydrogenases gene from Zymomonas mobilis CICC 41465 (AdhB) were transformed into Escherichia coli for higher alcohol production. SDS-PAGE results showed these two proteins were expressed in the recombinant strains. The resulting strain was incubated in LB medium at 37 °C in Erlenmeyer flasks and much more 3-methyl-1-butanol (104 mg/L) than isobutanol (24 mg/L) was produced. However, in 5 g/L glucose-containing medium, the production of two alcohols was similar, 156 and 161 mg/L for C4 (isobutanol) and C5 (3-methyl-1-butanol) alcohol, respectively. Effects of fermentation factors including temperature, glucose content, and α-keto acid on alcohol production were also investigated. The increase of glucose content and the adding of α-keto acids facilitated the production of C4 and C5 alcohols. The enzyme activities of pure Kivd on α-ketoisovalerate and α-ketoisocaproate were 26.77 and 21.24 μmol min−1 mg−1, respectively. Due to its ability on decarboxylation of α-ketoisovalerate and α-ketoisocaproate, the recombinant E. coli strain showed potential application on isoamyl alcohol and isobutanol production.






Similar content being viewed by others
References
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–90
Baez A, Cho KM, Liao JC (2011) High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal. Appl Microbiol Biotechnol 90:1681–1690
Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ (2011) Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol 77:3300–3310
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chen X, Nielsen KF, Borodina I, Kielland-Brandt MC, Karhumaa K (2011) Increased isobutanol production in Saccharomyces cerevisiae by overexpression of genes in valine metabolism. Biotechnol Biofuels 4:21–32
Connor MR, Liao JC (2008) Engineering of an Escherichia coli strain for the production of 3-methyl-1-butanol. Appl Environ Microbiol 74:5769–5775
de la Plaza M, de Palencia PF, Peláez C, Requena T (2004) Biochemical and molecular characterization of a-ketoisovalerate decarboxylase, an enzyme involved in the formation of aldehydes from amino acids by Lactococcus lactis. FEMS Microbiol Lett 238:367–374
Gocke D, Nguyen CL, Pohl M, Stillger T, Walter L, Müller M (2007) Branched-chain keto acid decarboxylase from Lactococcus lactis (KdcA), a valuable thiamine diphosphate-dependent enzyme for asymmetric C–C bond formation. Adv Synth Catal 349:1425–1435
Gogerty DS, Bobik TA (2010) Formation of isobutene from 3-hydroxy-3-methylbutyrate by diphosphomevalonate decarboxylase. Appl Environ Microbiol 76:8004–8010
Hazelwood LA, Daran JM, van Maris AJ, Pronk JT, Dickinson JR (2008) The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol 74:2259–2266
Kneen MM, Stan R, Yep A, Tyler RP, Saehuan C, McLeish MJ (2011) Characterization of a thiamin diphosphate-dependent phenylpyruvate decarboxylase from Saccharomyces cerevisiae. FEBS J 278:1842–1853
Koga J (1995) Structure and function of indolepyruvate decarboxylase, a key enzyme in indole-3-acetic acid biosynthesis. Biochem Biophys Acta 1(249):1–13
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lan EI, Liao JC (2013) Microbial synthesis of n-butanol, isobutanol, and other higher alcohols from diverse resources. Bioresour Technol 135:339–349
Larroy C, Rosario Fernandez M, Gonzalez E, Parés X, Biosca JA (2003) Properties and functional significance of Saccharomyces cerevisiae ADHVI. Chem Biol Interact 143–144:229–238
Li S, Wen J, Jia X (2011) Engineering Bacillus subtilis for isobutanol production by heterologous Ehrlich pathway construction and the biosynthetic 2-ketoisovalerate precursor pathway overexpression. Appl Microbiol Biotechnol 91:577–589
Lu J, Brigham CJ, Gai CS, Sinskey AJ (2012) Studies on the production of branched-chain alcohols in engineered Ralstonia eutropha. Appl Microbiol Biotechnol 96:283–297
Macho V, Kralik M, Jurecekova E, Hudec J, Jurecek L (2001) Dehydration of C-4 alkanols conjugated with a positional and skeletal isomerisation of the formed C-4 alkenes. Appl Catal A Gen 214:251–257
Oku H, Kaneda T (1988) Biosynthesis of branched-chain fatty acids in Bacillus subtilis. A decarboxylase is essential for branched-chain fatty acid synthetase. J Biol Chem 263:18386–18396
Palaniappan C, Sharma V, Hudspeth ME, Meganathan R (1992) Menaquinone (vitamin K2) biosynthesis: evidence that the Escherichia coli menD gene encodes both 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic acid synthase and alpha-ketoglutarate decarboxylase activities. J Bacteriol 174:8111–8118
Singer TP, Pensky J (1952) Isolation and properties of the a-carboxylase of wheat germ. J Biol Chem 196:375–388
Smit BA, van Hylckama Vlieg JET, Engels WJM, Meijer L, Wouters JT, Smit G (2005) Identification, cloning, and characterization of a Lactococcus lactis branched-chain α-keto acid decarboxylase involved in flavor formation. Appl Environ Microbiol 71:303–311
Smith KM, Cho KM, Liao JC (2010) Engineering Corynebacterium glutamicum for isobutanol production. Appl Microbiol Biotechnol 87:1045–1055
Vuralhan Z, Morais MA, Tai SL, Piper MD, Pronk JT (2003) Identification and characterization of phenylpyruvate decarboxylase genes in Saccharomyces cerevisiae. Appl Environ Microbiol 69:4534–4541
Wei J, Timler JG, Knutson CM, Barney BM (2013) Branched-chain 2-keto acid decarboxylases derived from Psychrobacter. FEMS Microbiol Lett 346:105–112
Zhang K, Sawaya MR, Eisenberg DS, Liao JC (2008) Expanding metabolism for biosynthesis of nonnatural alcohols. Proc Natl Acad Sci USA 105:20653–20658
Acknowledgments
This research was financially supported by National Natural Science Foundation of China (No. 2117623 and No. 21211140237), National High Technology Research and Development Program of China (863 Program) (2013AA065803), Guangdong science and technology research program (2013B010403021), Guangzhou science and technology research program (2013J4300026) and Foundation for Innovative Research of Guangzhou Institute of Energy Conversion, CAS (y407p11001).
Author information
Authors and Affiliations
Corresponding author
Additional information
X. Chen and J. Xu contribute equally to this work.
Rights and permissions
About this article
Cite this article
Chen, X., Xu, J., Yang, L. et al. Production of C4 and C5 branched-chain alcohols by engineered Escherichia. coli . J Ind Microbiol Biotechnol 42, 1473–1479 (2015). https://doi.org/10.1007/s10295-015-1656-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-015-1656-z