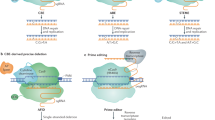
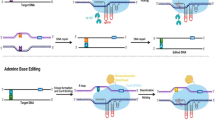
Abstract
The CRISPR/Cas systems have emerged as transformative tools for precisely manipulating plant genomes and enhancement. It has provided unparalleled applications from modifying the plant genomes to resistant enhancement. This review manuscript summarises the mechanism, application, and current challenges in the CRISPR/Cas genome editing technology. It addresses the molecular mechanisms of different Cas genes, elucidating their applications in various plants through crop improvement, disease resistance, and trait improvement. The advent of the CRISPR/Cas systems has enabled researchers to precisely modify plant genomes through gene knockouts, knock-ins, and gene expression modulation. Despite these successes, the CRISPR/Cas technology faces challenges, including off-target effects, Cas toxicity, and efficiency. In this manuscript, we also discuss these challenges and outline ongoing strategies employed to overcome these challenges, including the development of novel CRISPR/Cas variants with improved specificity and specific delivery methods for different plant species. The manuscript will conclude by addressing the future perspectives of the CRISPR/Cas technology in plants. Although this review manuscript is not conclusive, it aims to provide immense insights into the current state and future potential of CRISPR/Cas in sustainable and secure plant production.


Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
References
Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F (2017) RNA targeting with CRISPR–Cas13. Nature 550(7675):280–284. https://doi.org/10.1038/nature24049
Abudayyeh OO, Gootenberg JS, Franklin B, Koob J, Kellner MJ, Ladha A, Joung J, Kirchgatterer P, Cox DB, Zhang F (2019) A cytosine deaminase for programmable single-base RNA editing. Science 365(6451):382–386
Adeyinka OS, Tabassum B, Koloko BL, Ogungbe IV (2023) Enhancing the quality of staple food crops through CRISPR/Cas-mediated site-directed mutagenesis. Planta 257(4):78. https://doi.org/10.1007/s00425-023-04110-6
Ahmar S, Hensel G, Gruszka D (2023) CRISPR/Cas9-mediated genome editing techniques and new breeding strategies in cereals–current status, improvements, and perspectives. Biotechnol Adv 69:108248. https://doi.org/10.1016/j.biotechadv.2023.108248
Alberti F, Leng DJ, Wilkening I, Song L, Tosin M, Corre C (2019) Triggering the expression of a silent gene cluster from genetically intractable bacteria results in scleric acid discovery. Chemical Science 10(2):453–463. https://doi.org/10.1039/C8SC03814G
Aman R, Ali Z, Butt H, Mahas A, Aljedaani F, Khan MZ, Ding S, Mahfouz M (2018a) RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol 19(1):1. https://doi.org/10.1186/s13059-017-1381-1
Aman R, Mahas A, Butt H, Ali Z, Aljedaani F, Mahfouz M (2018b) Engineering RNA virus interference via the CRISPR/Cas13 machinery in Arabidopsis. Viruses 10(12):732
Anantharaman V, Makarova KS, Burroughs AM, Koonin EV, Aravind L (2013) Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol Direct 8(1):1–28
Ashraf S, Ahmad A, Khan SH, Jamil A, Sadia B, Brown JK (2023) LbCas12a mediated suppression of Cotton leaf curl Multan virus. Front Plant Sci 14. https://doi.org/10.3389/fpls.2023.1233295
Awan MJA, Amin I, Mansoor S (2022) CRISPR-Cas12c: a noncleaving DNA binder with minimal PAM requirement. Trends Biotechnol 40(10):1141–1143. https://doi.org/10.1016/j.tibtech.2022.07.005
Basu U, Riaz Ahmed S, Bhat BA, Anwar Z, Ali A, Ijaz A, Gulzar A, Bibi A, Tyagi A, Nebapure SM, Goud CA, Ahanger SA, Ali S, Mushtaq M (2023) A CRISPR way for accelerating cereal crop improvement: Progress and challenges [Review]. Front Genet 13:866976. https://doi.org/10.3389/fgene.2022.866976
Biswas P, Ghorai M, Nandy S, Nongdam P, Pandey DK, Dwivedi P, Shekhawat MS, Dey A (2023) Application of CRISPR/Cas system in optimizing nutrients and anti-nutrients content in fruits. Vegetos:1–9. https://doi.org/10.1007/s42535-023-00652-y
Bo W, Zhaohui Z, Huanhuan Z, Xia W, Binglin L, Lijia Y, Xiangyan H, Deshui Y, Xuelian Z, Chunguo W, Wenqin S, Chengbin C, Yong Z (2019) Targeted Mutagenesis of NAC Transcription Factor Gene, OsNAC041, Leading to Salt Sensitivity in Rice. Rice Science 26(2):98–108. https://doi.org/10.1016/j.rsci.2018.12.005
Buchman AB, Brogan DJ, Sun R, Yang T, Hsu PD, Akbari OS (2020) Programmable RNA targeting using CasRx in flies. The CRISPR Journal 3(3):164–176
Butt H, Eid A, Ali Z, Atia MA, Mokhtar MM, Hassan N, Lee CM, Bao G, Mahfouz MM (2017) Efficient CRISPR/Cas9-mediated genome editing using a chimeric single-guide RNA molecule. Front Plant Sci 8:1441
Bykonya AG, Lavrov AV, Smirnikhina SA (2023) Methods for CRISPR-Cas as Ribonucleoprotein Complex Delivery In Vivo. Mol Biotechnol 65(2):181–195. https://doi.org/10.1007/s12033-022-00479-z
Campa M, Miranda S, Licciardello C, Lashbrooke JG, Dalla Costa L, Guan Q, Spök A, Malnoy M (2023) Application of new breeding techniques in fruit trees Plant Physiol 3:kiad374. https://doi.org/10.1093/plphys/kiad374
Cao G, Dong J, Chen X, Lu P, Xiong Y, Peng L, Li J, Huo D, Hou C (2022) Simultaneous detection of CaMV35S and T-nos utilizing CRISPR/Cas12a and Cas13a with multiplex-PCR (MPT-Cas12a/13a). Chem Commun 58(43):6328–6331
Carroll D (2011) Genome engineering with zinc-finger nucleases. Genetics 188(4):773–782
Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA (2018a) CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360(6387):436–439
Chen R, Xu Q, Liu Y, Zhang J, Ren D, Wang G, Liu Y (2018b) Generation of transgene-free maize male sterile lines using the CRISPR/Cas9 system. Front Plant Sci 9:1180
Cheng X, Li Z, Shan R, Li Z, Wang S, Zhao W, Zhang H, Chao L, Peng J, Fei T, Li W (2023) Modeling CRISPR-Cas13d on-target and off-target effects using machine learning approaches. Nat Commun 14(1):752. https://doi.org/10.1038/s41467-023-36316-3
Chylinski K, Le Rhun A, Charpentier E (2013) The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol 10(5):726–737
Cox DB, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F (2017) RNA editing with CRISPR-Cas13. Science 358(6366):1019–1027
Das T, Anand U, Pal T, Mandal S, Kumar M, Radha Gopalakrishnan AV, Lastra JMPdl, Dey A (2023) Exploring the potential of CRISPR/Cas genome editing for vegetable crop improvement: An overview of challenges and approaches. Biotechnol Bioeng 120(5):6894. https://doi.org/10.1002/bit.28344
Deng X, Osikpa E, Yang J, Oladeji SJ, Smith J, Gao X, Gao Y (2023) Structural basis for the activation of a compact CRISPR-Cas13 nuclease. Nat Commun 14(1):5845
Dong D, Ren K, Qiu X, Zheng J, Guo M, Guan X, Liu H, Li N, Zhang B, Yang D (2016) The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 532(7600):522–526
Dong L, Li L, Liu C, Liu C, Geng S, Li X, Huang C, Mao L, Chen S, Xie C (2018) Genome editing and double-fluorescence proteins enable robust maternal haploid induction and identification in maize. Mol Plant 11(9):1214–1217
Dong OX, Yu S, Jain R, Zhang N, Duong PQ, Butler C, Li Y, Lipzen A, Martin JA, Barry KW, Schmutz J, Tian L, Ronald PC (2020) Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat Commun 11(1):1178. https://doi.org/10.1038/s41467-020-14981-y
Duan Y-B, Li J, Qin R-Y, Xu R-F, Li H, Yang Y-C, Ma H, Li L, Wei P-C, Yang J-B (2016) Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol Biol 90(1):49–62. https://doi.org/10.1007/s11103-015-0393-z
East-Seletsky A, O’Connell MR, Burstein D, Knott GJ, Doudna JA (2017) RNA targeting by functionally orthogonal type VI-A CRISPR-Cas enzymes. Molecular cell 66(3):373-383.e373
Fan Y, Xin S, Dai X, Yang X, Huang H, Hua Y (2020) Efficient genome editing of rubber tree (hevea brasiliensis) protoplasts using CRISPR/Cas9 ribonucleoproteins. Industrial Crops and Products 146:112146. https://doi.org/10.1016/j.indcrop.2020.112146
Fan S, Zhang L, Tang M, Cai Y, Liu J, Liu H, Liu J, Terzaghi W, Wang H, Hua W, Zheng M (2021) CRISPR/Cas9-targeted mutagenesis of the BnaA03.BP gene confers semi-dwarf and compact architecture to rapeseed (Brassica napus L.). Plant Biotechnol J 19(12):2383–2385. https://doi.org/10.1111/pbi.13703
Fatollahi Arani S, Zeinoddini M (2023) Gene editing: biosecurity challenges and risks. Journal of Police Medicine 12(1):1–19
Feng W, Peng H, Xu J, Liu Y, Pabbaraju K, Tipples G, Joyce MA, Saffran HA, Tyrrell DL, Babiuk S (2021) Integrating reverse transcription recombinase polymerase amplification with CRISPR technology for the one-tube assay of RNA. Anal Chem 93(37):12808–12816
Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E (2016) The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532(7600):517–521
Gao J, Luo T, Lin N, Zhang S, Wang J (2020) A new tool for CRISPR-Cas13a-based cancer gene therapy. Molecular Therapy-Oncolytics 19:79–92
Géry C, Téoulé E (2022) Cold acclimation diversity in Arabidopsis thaliana: CRISPR/Cas9 as a tool to fine analysis of Tandem Gene Arrays, application to CBF genes. Dev Genes Evol 232(5):147–154. https://doi.org/10.1007/s00427-022-00693-4
González MN, Massa GA, Andersson M, Turesson H, Olsson N, Fält A-S, Storani L, Décima Oneto CA, Hofvander P, Feingold SE (2020) Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front Plant Sci 10:1649
González MN, Massa GA, Andersson M, DécimaOneto CA, Turesson H, Storani L, Olsson N, Fält A-S, Hofvander P, Feingold SE (2021) Comparative potato genome editing: Agrobacterium tumefaciens-mediated transformation and protoplasts transfection delivery of CRISPR/Cas9 components directed to StPPO2 gene. Plant Cell, Tissue and Organ Culture (PCTOC) 145(2):291–305. https://doi.org/10.1007/s11240-020-02008-9
Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F (2017) Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356(6336):438–442. https://doi.org/10.1126/science.aam9321
Gunitseva N, Evteeva M, Borisova A, Patrushev M, Subach F (2023) RNA-dependent RNA targeting by CRISPR-Cas systems: characterizations and applications. Int J Mol Sci 24(8):6894. https://doi.org/10.3390/ijms24086894
Gupta D, Bhattacharjee O, Mandal D, Sen MK, Dey D, Dasgupta A, Kazi TA, Gupta R, Sinharoy S, Acharya K (2019) CRISPR-Cas9 system: A new-fangled dawn in gene editing. Life Sci 232:116636
Gurel F, Wu Y, Pan C, Cheng Y, Li G, Zhang T, Qi Y (2023) On-and Off-Target Analyses of CRISPR-Cas12b Genome Editing Systems in Rice. The CRISPR Journal 6(1):62–74
Haft DH, Selengut J, Mongodin EF, Nelson KE (2005) A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1(6):e60
Hajiahmadi Z, Movahedi A, Wei H, Li D, Orooji Y, Ruan H, Zhuge Q (2019) Strategies to increase on-target and reduce off-target effects of the CRISPR/Cas9 system in plants. Int J Mol Sci 20(15):3719. https://doi.org/10.3390/ijms20153719
Hamdan MF, Karlson CKS, Teoh EY, Lau SE, Tan BC (2022) Genome editing for sustainable crop improvement and mitigation of biotic and abiotic stresses. Plants (Basel) 11(19):2625. https://doi.org/10.3390/plants11192625
Han X, Chen Z, Li P, Xu H, Liu K, Zha W, Li S, Chen J, Yang G, Huang J (2022) Development of novel rice germplasm for salt-tolerance at seedling stage using CRISPR-Cas9. Sustainability 14(5):2621
Han J, Li X, Li W, Yang Q, Li Z, Cheng Z, Lv L, Zhang L, Han D (2023) Isolation and preliminary functional analysis of FvICE1, involved in cold and drought tolerance in Fragaria vesca through overexpression and CRISPR/Cas9 technologies. Plant Physiology and Biochemistry 196:270–280. https://doi.org/10.1016/j.plaphy.2023.01.048
Hanlon KS, Kleinstiver BP, Garcia SP, Zaborowski MP, Volak A, Spirig SE, Muller A, Sousa AA, Tsai SQ, Bengtsson NE (2019) High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat Commun 10(1):4439
Hillary VE, Ceasar SA (2022) A review on the mechanism and applications of CRISPR/Cas9/Cas12/Cas13/Cas14 proteins utilized for genome engineering. Mol Biotechnol 65(3):311–325. https://doi.org/10.1007/s12033-022-00567-0
Hillmann K (2019) Looking for maize genes involved in cold response: producing knockouts for arabidopsis homologs of maize candidate genes using a CRISPR/Cas9 Approach. Departmental Honors Projects 82. https://digitalcommons.hamline.edu/dhp/82
Hochstrasser ML, Doudna JA (2015) Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends in Biochemical Sciences 40(1):58–66. https://doi.org/10.1016/j.tibs.2014.10.007
Hu X, Yang J, Cheng C, Zhou J, Niu F, Wang X, Zhang M, Cao L, Chu H (2018) Targeted editing of rice SD1 gene using CRISPR/Cas9 system. Chin J Rice Sci 32(3):219–225
Hua K, Tao X, Yuan F, Wang D, Zhu J-K (2018) Precise A· T to G· C base editing in the rice genome. Mol Plant 11(4):627–630
Hwang S, Maxwell KL (2023) Diverse mechanisms of CRISPR-Cas9 inhibition by type II anti-CRISPR proteins. J Mol Biol435(7):168041. https://doi.org/10.1016/j.jmb.2023.168041
Hwarari D, Guan Y, Ahmad B, Movahedi A, Min T, Hao Z, Lu Y, Chen J, Yang L (2022) ICE-CBF-COR signaling cascade and its regulation in plants responding to cold stress. Int J Mol Sci 23(3):1549
Illouz-Eliaz N, Nissan I, Nir I, Ramon U, Shohat H, Weiss D (2020) Mutations in the tomato gibberellin receptors suppress xylem proliferation and reduce water loss under water-deficit conditions. J Exp Bot 71(12):3603–3612. https://doi.org/10.1093/jxb/eraa137
Jia H, Orbović V, Wang N (2019) CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol J 17(10):1928–1937
Karmakar S, Das P, Panda D, Xie K, Baig MJ, Molla KA (2022) A detailed landscape of CRISPR-Cas-mediated plant disease and pest management. Plant Science 323:111376. https://doi.org/10.1016/j.plantsci.2022.111376
Kaul T, Thangaraj A, Jain R, Bharti J, Kaul R, Verma R, Sony SK, Motelb KFA, Yadav P, Agrawal PK (2024) CRISPR/Cas9-mediated homology donor repair base editing system to confer herbicide resistance in maize (Zea mays L.). Plant Physiol Biochem 207:108374. https://doi.org/10.1016/j.plaphy.2024.108374
Kavuri NR, Ramasamy M, Qi Y, Mandadi K (2022) Applications of CRISPR/Cas13-Based RNA Editing in Plants. Cells 11(17):2665
Khan Z, Ali Z, Khan AA, Sattar T, Zeshan A, Saboor T, Binyamin B (2022) History and classification of CRISPR/Cas system. In: The CRISPR/Cas Tool Kit for Genome Editing. Springer, pp 29–52. https://doi.org/10.1007/978-981-16-6305-5_2
Kharbikar L, Konwarh R, Chakraborty M, Nandanwar S, Marathe A, Yele Y, Ghosh PK, Sanan-Mishra N, Singh AP (2023) 3Bs of CRISPR-Cas mediated genome editing in plants: exploring the basics, bioinformatics and biosafety landscape. Physiol Mol Biol Plants 29(12):1825–1850. https://doi.org/10.1007/s12298-023-01397-3
Knott GJ, East-Seletsky A, Cofsky JC, Holton JM, Charles E, O’Connell MR, Doudna JA (2017) Guide-bound structures of an RNA-targeting A-cleaving CRISPR–Cas13a enzyme. Nat Struct Mol Biol 24(10):825–833. https://doi.org/10.1038/nsmb.3466
Kocsisova Z, Coneva V (2023) Strategies for delivery of CRISPR/Cas-mediated genome editing to obtain edited plants directly without transgene integration. Front Genome Ed 5:1209586. https://doi.org/10.3389/fgeed.2023.1209586
Laforest LC, Nadakuduti SS (2022) Advances in delivery mechanisms of CRISPR gene-editing reagents in plants. Frontiers in Genome Editing 4:830178
Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q, Zhang R, Gao C (2018a) Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol 19:1–9
Li R, Zhang L, Wang L, Chen L, Zhao R, Sheng J, Shen L (2018b) Reduction of tomato-plant chilling tolerance by CRISPR–Cas9-mediated SlCBF1 mutagenesis. J Agric Food Chem 66(34):9042–9051
Li B, Rui H, Li Y, Wang Q, Alariqi M, Qin L, Sun L, Ding X, Wang F, Zou J, Wang Y, Yuan D, Zhang X, Jin S (2019a) Robust CRISPR/Cpf1 (Cas12a)-mediated genome editing in allotetraploid cotton (Gossypium hirsutum). Plant Biotechnol J 17(10):1862–1864. https://doi.org/10.1111/pbi.13147
Li R, Liu C, Zhao R, Wang L, Chen L, Yu W, Zhang S, Sheng J, Shen L (2019b) CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol 19(1):38. https://doi.org/10.1186/s12870-018-1627-4
Li C, Zhang R, Meng X, Chen S, Zong Y, Lu C, Qiu J-L, Chen Y-H, Li J, Gao C (2020a) Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat Biotechnol 38(7):875–882. https://doi.org/10.1038/s41587-019-0393-7
Li J, Wang Z, He G, Ma L, Deng XW (2020b) CRISPR/Cas9-mediated disruption of TaNP1 genes results in complete male sterility in bread wheat. Journal of Genetics and Genomics 47(5):263–272. https://doi.org/10.1016/j.jgg.2020.05.004
Li C, Chu W, Gill RA, Sang S, Shi Y, Hu X, Yang Y, Zaman QU, Zhang B (2022a) Computational Tools and Resources for CRISPR/Cas Genome Editing. Genomics Proteomics Bioinformatics 21(1):108–126. https://doi.org/10.1016/j.gpb.2022.02.006
Li S, Lin D, Zhang Y, Deng M, Chen Y, Lv B, Li B, Lei Y, Wang Y, Zhao L, Liang Y, Liu J, Chen K, Liu Z, Xiao J, Qiu J-L, Gao C (2022b) Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602(7897):455–460. https://doi.org/10.1038/s41586-022-04395-9
Li G, Zhang Y, Dailey M, Qi Y (2023a) Hs1Cas12a and Ev1Cas12a confer efficient genome editing in plants. Front Genome Ed 5:1251903. https://doi.org/10.3389/fgeed.2023.1251903
Li Z, Zhong Z, Wu Z, Pausch P, Al-Shayeb B, Amerasekera J, Doudna JA, Jacobsen SE (2023b) Genome editing in plants using the compact editor CasΦ. Proc Natl Acad Sci U S A 120(4):e2216822120. https://doi.org/10.1073/pnas.2216822120
Li X, Li H, Zhao Y, Zong P, Zhan Z, Piao Z (2020c) Establishment of Agrobacterium-mediated genetic transformation and application of CRISPR/Cas9 gene-editing system to Chinese cabbage (Brassica rapa L. ssp. pekinensis)
Liu L, Li X, Ma J, Li Z, You L, Wang J, Wang M, Zhang X, Wang Y (2017a) The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell 170(4):714-726.e710
Liu L, Li X, Wang J, Wang M, Chen P, Yin M, Li J, Sheng G, Wang Y (2017b) Two distant catalytic sites are responsible for C2c2 RNase activities. Cell 168(1–2):121-134.e112
Liu L, Zhang J, Xu J, Li Y, Guo L, Wang Z, Zhang X, Zhao B, Guo Y-D, Zhang N (2020) CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Science 301:110683. https://doi.org/10.1016/j.plantsci.2020.110683
Liu X, Zhang S, Jiang Y, Yan T, Fang C, Hou Q, Wu S, Xie K, An X, Wan X (2022) Use of CRISPR/Cas9-based gene editing to simultaneously mutate multiple homologous genes required for pollen development and male fertility in maize. Cells 11(3):439
Liu Q, Zhao C, Sun K, Deng Y, Li Z (2023) Engineered biocontainable RNA virus vectors for non-transgenic genome editing across crop species and genotypes. Mol Plant. https://doi.org/10.1016/j.molp.2023.02.003
Liyanage DW, Yevtushenko DP, Konschuh M, Bizimungu B, Lu Z-X (2021) Processing strategies to decrease acrylamide formation, reducing sugars and free asparagine content in potato chips from three commercial cultivars. Food Control 119:107452
López-Casado G, Sánchez-Raya C, Ric-Varas PD, Paniagua C, Blanco-Portales R, Muñoz-Blanco J, Pose S, Matas AJ, Mercado JA (2023) CRISPR/Cas9 editing of the polygalacturonase FaPG1 gene improves strawberry fruit firmness. Hortic Res 10(3):uhad011. https://doi.org/10.1093/hr/uhad011
Loureiro A, da Silva GJ (2019) Crispr-cas: Converting a bacterial defence mechanism into a state-of-the-art genetic manipulation tool. Antibiotics 8(1):18
Lv P, Su F, Chen F, Yan C, Xia D, Sun H, Li S, Duan Z, Ma C, Zhang H (2023) Genome editing in rice using CRISPR/Cas12i3. Plant Biotechnol J 22(2):379–385. https://doi.org/10.1111/pbi.14192
Ly DNP, Iqbal S, Fosu-Nyarko J, Milroy S, Jones MG (2023) Multiplex CRISPR-Cas9 Gene-Editing Can Deliver Potato Cultivars with Reduced Browning and Acrylamide. Plants 12(2):379
Ma X, Zhang X, Liu H, Li Z (2020) Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nature Plants 6(7):773–779. https://doi.org/10.1038/s41477-020-0704-5
Mahas A, Aman R, Mahfouz M (2019) CRISPR-Cas13d mediates robust RNA virus interference in plants. Genome Biol 20(1):263. https://doi.org/10.1186/s13059-019-1881-2
Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJ, Charpentier E, Cheng D, Haft DH, Horvath P (2020) Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18(2):67–83
Makarova KS, Wolf YI, Koonin EV (2022) Evolutionary classification of CRISPR‐Cas systems. Crispr: Biology and Applications, pp 13–38. https://doi.org/10.1002/9781683673798.ch2
Meeske AJ, Nakandakari-Higa S, Marraffini LA (2019) Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature 570(7760):241–245
Min T, Hwarari D, Li D, Movahedi A, Yang L (2022) CRISPR-based genome editing and its applications in woody plants. Int J Mol Sci 23(17):10175
Minh BM, Hanh HH, Hue HTT (2020) Construction of CRISPR/Cas9 expression vectors habouring gRNA targeted on SlIAA9 gene of tomato. Vietnam Journal of Biotechnology 18(1):147–156
Mubarik MS, Khan SH, Sajjad M (2021) Key applications of CRISPR/Cas for yield and nutritional improvement. In: Ahmad A, Khan SH, Khan Z (eds) CRISPR Crops: The Future of Food Security Springer, Singapore, pp 213–230. https://doi.org/10.1007/978-981-15-7142-8_7
Naeem M, Alkhnbashi OS (2023) Current Bioinformatics Tools to Optimize CRISPR/Cas9 Experiments to Reduce Off-Target Effects. Int J Mol Sci 24(7):6261
Newsom S, Parameshwaran HP, Martin L, Rajan R (2021) The CRISPR-Cas mechanism for adaptive immunity and alternate bacterial functions fuels diverse biotechnologies. Front Cell Infect Microbiol 10:619763
Nguyen NH, Bui TP, Le NT, Nguyen CX, Le MTT, Dao NT, Phan Q, Van Le T, To HMT, Pham NB (2023) Disrupting Sc-uORFs of a transcription factor bZIP1 using CRISPR/Cas9 enhances sugar and amino acid contents in tomato (Solanum lycopersicum). Planta 257(3):57
O’Connell MR (2019) Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR–Cas systems. J Mol Biol 431(1):66–87
Ogata T, Ishizaki T, Fujita M, Fujita Y (2020) CRISPR/Cas9-targeted mutagenesis of OsERA1 confers enhanced responses to abscisic acid and drought stress and increased primary root growth under nonstressed conditions in rice. PLoS ONE 15(12):e0243376
Okada A, Arndell T, Borisjuk N, Sharma N, Watson-Haigh NS, Tucker EJ, Baumann U, Langridge P, Whitford R (2019) CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol J 17(10):1905–1913
Ozyigit II (2020) Gene transfer to plants by electroporation: methods and applications. Mol Biol Rep 47(4):3195–3210. https://doi.org/10.1007/s11033-020-05343-4
Parent B (2023) Learning from Icarus: The impact of CRISPR on gene editing ethics. In: Handbook of Bioethical Decisions. Volume I: Decisions at the Bench. Springer 2:9–27. https://doi.org/10.1007/978-3-031-29451-8_2
Pausch P, Al-Shayeb B, Bisom-Rapp E, Tsuchida CA, Li Z, Cress BF, Knott GJ, Jacobsen SE, Banfield JF, Doudna JA (2020) CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 369(6501):333–337. https://doi.org/10.1126/science.abb1400
Peng X, Ma X, Lu S, Li Z (2020) A Versatile Plant Rhabdovirus-Based Vector for Gene Silencing, miRNA Expression and Depletion, and Antibody Production. Front Plant Sci 11:627880. https://doi.org/10.3389/fpls.2020.627880
Pramanik D, Shelake RM, Park J, Kim MJ, Hwang I, Park Y, Kim J-Y (2021) CRISPR/Cas9-mediated generation of pathogen-resistant tomato against tomato yellow leaf curl virus and powdery mildew. Int J Mol Sci 22(4):1878
Pu Y, Liu C, Li J, Aerzu GT, Hu Y, Liu X (2018) Different SlU6 promoters cloning and establishment of CRISPR/Cas9 mediated gene editing system in tomato. Scientia Agricultura Sinica 51(2):315–326
Que Z, Lu Q, Liu T, Li S, Zou J, Chen G (2023) The rice annexin gene OsAnn5 is a positive regulator of cold stress tolerance at the seedling stage. Plant Direct 7(11):e539. https://doi.org/10.1007/978-3-031-29451-8_2
Ran FA, Hsu PD, Lin C-Y, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154(6):1380–1389
Ribeiro JA, Varanda C, Materatski P, Campos MD, Albuquerque A, Patanita M, Osa N, Félix M (2023) CRISPR/Cas13 system: a technology for the successful control of plant viruses. Book of abstracts: Science Changing Policys, p 101. http://hdl.handle.net/10400.15/4686
Saeed S, Usman B, Shim S-H, Khan SU, Nizamuddin S, Saeed S, Shoaib Y, Jeon J-S, Jung K-H (2022) CRISPR/Cas-mediated editing of cis-regulatory elements for crop improvement. Plant Sci 324:111435
Santillán Martínez MI, Bracuto V, Koseoglou E, Appiano M, Jacobsen E, Visser RG, Wolters A-MA, Bai Y (2020) CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol 20(1):1–13
Santosh Kumar VV, Verma RK, Yadav SK, Yadav P, Watts A, Rao MV, Chinnusamy V (2020) CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol Mol Biol Plants 26(6):1099–1110. https://doi.org/10.1007/s12298-020-00819-w
Saraswat P, Ranjan R (2022) CRISPR-Cas system: a precise tool for plant genome editing. The Nucleus 65(1):81–98. https://doi.org/10.1007/s13237-021-00353-4
Schulze-Lefert P, Vogel J (2000) Closing the ranks to attack by powdery mildew. Trends Plant Sci 5(8):343–348
Sharma P, Pandey A, Malviya R, Dey S, Karmakar S, Gayen D (2023) Genome editing for improving nutritional quality, post-harvest shelf life and stress tolerance of fruits, vegetables, and ornamentals. Frontiers in Genome Editing 5:1094965
Shen C, Que Z, Xia Y, Tang N, Li D, He R, Cao M (2017) Knock out of the annexin gene OsAnn3 via CRISPR/Cas9-mediated genome editing decreased cold tolerance in rice. Journal of Plant Biology 60:539–547
Shkryl Y, Yugay Y, Avramenko T, Grigorchuk V, Gorpenchenko T, Grischenko O, Bulgakov V (2021) CRISPR/Cas9-mediated knockout of HOS1 reveals its role in the regulation of secondary metabolism in Arabidopsis thaliana. Plants 10(1):104
Shmakov SA, Faure G, Makarova KS, Wolf YI, Severinov KV, Koonin EV (2019) Systematic prediction of functionally linked genes in bacterial and archaeal genomes. Nat Protoc 14(10):3013–3031
Singer SD, Burton Hughes K, Subedi U, Dhariwal GK, Kader K, Acharya S, Chen G, Hannoufa A (2022) The CRISPR/Cas9-mediated modulation of SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE 8 in alfalfa leads to distinct phenotypic outcomes. Front Plant Sci 12:3203
Smargon AA, Cox DB, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, Abudayyeh OA, Essletzbichler P, Shmakov S, Makarova KS (2017) Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Molecular cell 65(4):618-630.e617
Subburaj S, Chung SJ, Lee C, Ryu S-M, Kim DH, Kim J-S, Bae S, Lee G-J (2016) Site-directed mutagenesis in Petunia × hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep 35(7):1535–1544. https://doi.org/10.1007/s00299-016-1937-7
Tang H, Liu H, Zhou Y, Liu H, Du L, Wang K, Ye X (2021) Fertility recovery of wheat male sterility controlled by Ms2 using CRISPR/Cas9. Plant Biotechnol J 19(2):224
Tashkandi M, Ali Z, Aljedaani F, Shami A, Mahfouz MM (2018) Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal Behav 13(10):e1525996
Thapliyal G, Bhandari MS, Vemanna RS, Pandey S, Meena RK, Barthwal S (2023) Engineering traits through CRISPR/cas genome editing in woody species to improve forest diversity and yield. Crit Rev Biotechnol 43(6):884–903
Tong Y, Whitford CM, Blin K, Jørgensen TS, Weber T, Lee SY (2020) CRISPR–Cas9, CRISPRi and CRISPR-BEST-mediated genetic manipulation in streptomycetes. Nat Protoc 15(8):2470–2502. https://doi.org/10.1038/s41596-020-0339-z
Toufikuzzaman M, Samee MAH, Rahman MS (2023) CRISPR-DIPOFF: an interpretable deep learning approach for CRISPR Cas-9 off-target prediction. Brief Bioinform 25(2):bbad530. https://doi.org/10.1093/bib/bbad530
Tran MT, Doan DTH, Kim J, Song YJ, Sung YW, Das S, Kim EJ, Son GH, Kim SH, Van Vu T, Kim J-Y (2021) CRISPR/Cas9-based precise excision of SlHyPRP1 domain(s) to obtain salt stress-tolerant tomato. Plant Cell Rep 40(6):999–1011. https://doi.org/10.1007/s00299-020-02622-z
Usman B, Nawaz G, Zhao N, Liao S, Liu Y, Li R (2020) Precise editing of the OsPYL9 gene by RNA-guided Cas9 nuclease confers enhanced drought tolerance and grain yield in rice (Oryza sativa L.) by regulating circadian rhythm and abiotic stress responsive proteins. International Journal of Molecular Sciences 21(21):7854
Van Esse HP, Reuber TL, van der Does D (2020) Genetic modification to improve disease resistance in crops. New Phytol 225(1):70–86
Verma V, Kumar A, Partap M, Thakur M, Bhargava B (2023) CRISPR-Cas: A robust technology for enhancing consumer-preferred commercial traits in crops. Front Plant Sci 14:1122940
Wan D-Y, Guo Y, Cheng Y, Hu Y, Xiao S, Wang Y, Wen Y-Q (2020) CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic Res 7:11. https://doi.org/10.1038/s41438-020-0339-8
Wang F, Wang C, Liu P, Lei C, Hao W, Gao Y, Liu Y-G, Zhao K (2016a) Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 11(4):e0154027
Wang M, Zuris JA, Meng F, Rees H, Sun S, Deng P, Han Y, Gao X, Pouli D, Wu Q, Georgakoudi I, Liu DR, Xu Q (2016) Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proceedings of the National Academy of Sciences 113(11):2868–2873. https://doi.org/10.1073/pnas.1520244113
Wang K, Zhao Q-W, Liu Y-F, Sun C-F, Chen X-A, Burchmore R, Burgess K, Li Y-Q, Mao X-M (2019) Multi-layer controls of Cas9 activity coupled with ATP synthase over-expression for efficient genome editing in Streptomyces. Frontiers in Bioengineering and Biotechnology 7:304
Wang Q, Alariqi M, Wang F, Li B, Ding X, Rui H, Li Y, Xu Z, Qin L, Sun L, Li J, Zou J, Lindsey K, Zhang X, Jin S (2020) The application of a heat-inducible CRISPR/Cas12b (C2c1) genome editing system in tetraploid cotton (G. hirsutum) plants. Plant Biotechnol J 18(12):2436–2443. https://doi.org/10.1111/pbi.13417
Wang Q, Xie F, Tong Y, Habisch R, Yang B, Zhang L, Müller R, Fu C (2020b) Dual-function chromogenic screening-based CRISPR/Cas9 genome editing system for actinomycetes. Appl Microbiol Biotechnol 104(1):225–239. https://doi.org/10.1007/s00253-019-10223-4
Wang B, Zhang T, Yin J, Yu Y, Xu W, Ding J, Patel DJ, Yang H (2021) Structural basis for self-cleavage prevention by tag: anti-tag pairing complementarity in type VI Cas13 CRISPR systems. Molecular cell 81(5):1100-1115.e1105
Wang Z, Wong DCJ, Wang Y, Xu G, Ren C, Liu Y, Kuang Y, Fan P, Li S, Xin H (2021b) GRAS-domain transcription factor PAT1 regulates jasmonic acid biosynthesis in grape cold stress response. Plant Physiol 186(3):1660–1678
Wang W, Wang W, Pan Y, Tan C, Li H, Chen Y, Liu X, Wei J, Xu N, Han Y, Gu H, Ye R, Ding Q, Ma C (2022a) A new gain-of-function OsGS2/GRF4 allele generated by CRISPR/Cas9 genome editing increases rice grain size and yield. The Crop Journal 10(4):1207–1212. https://doi.org/10.1016/j.cj.2022.01.004
Wang H, Ai L, Xia Y, Wang G, Xiong Z, Song X (2024) Software-based screening for efficient sgRNAs in Lactococcus lactis. J Sci Food Agric 104(2):1200–1206
Wang X, Li D, Tan X, Cai C, Zhang X, Shen Z, Yang A, Fu X, Liu D (2022b) CRISPR/Cas9-mediated targeted mutagenesis of two homoeoalleles in tobacco confers resistance to powdery mildew. Euphytica 219(6):67. https://doi.org/10.1007/s10681-023-03196-z
Wei Y, Yang Z, Zong C, Wang B, Ge X, Tan X, Liu X, Tao Z, Wang P, Ma C (2021) Trans single-stranded DNA cleavage via CRISPR/Cas14a1 activated by target RNA without destruction. Angew Chem 60(45):24241–24247. https://doi.org/10.1002/anie.202110384
Wei T, Jiang L, You X, Ma P, Xi Z, Wang NN (2023) Generation of Herbicide-Resistant Soybean by Base Editing. Biology (Basel) 12(5):741. https://www.mdpi.com/2079-7737/12/5/741
Wenjing W, Chen Q, Singh PK, Huang Y, Pei D (2020) CRISPR/Cas9 edited HSFA6a and HSFA6b of Arabidopsis thaliana offers ABA and osmotic stress insensitivity by modulation of ROS homeostasis. Plant Signal Behav 15(12):1816321
Wolter F, Schindele P, Puchta H (2019) Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol 19(1):1–8
Wu F, Qiao X, Zhao Y, Zhang Z, Gao Y, Shi L, Du H, Wang L, Zhang YJ, Zhang Y, Liu L, Wang Q, Kong D (2020) Targeted mutagenesis in Arabidopsis thaliana using CRISPR-Cas12b/C2c1. J Integr Plant Biol 62(11):1653–1658. https://doi.org/10.1111/jipb.12944
Xu P, Wang H, Tu R, Liu Q, Wu W, Fu X, Cao L, Shen X (2019) Orientation improvement of blast resistance in rice via CRISPR/Cas9 system. Chin J Rice Sci 33(4):313–322
Xu W, Zhang C, Yang Y, Zhao S, Kang G, He X, Song J, Yang J (2020) Versatile nucleotides substitution in plant using an improved prime editing system. Mol Plant 13(5):675–678
Xu J, Liu Z, Zhang Z, Wu T (2023a) Unlocking the full potential of Cas12a: exploring the effects of substrate and reaction conditions on trans-cleavage activity. Anal Chem 95(28):10664–10669
Xu T, Yang X, Feng X, Luo H, Luo C, Jia M-a, Lei L (2023b) Sensitive and visual detection of brassica yellows virus using RT-LAMP-coupled CRISPR-Cas12 assay. Phytopathology(ja) 19:PHYTO06230195R. https://doi.org/10.1094/PHYTO-06-23-0195-R
Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, Scott DA (2018) Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Molecular cell 70(2):327-339.e325. https://doi.org/10.1016/j.molcel.2018.02.028
Yan D, Ren B, Liu L, Yan F, Li S, Wang G, Sun W, Zhou X, Zhou H (2021) High-efficiency and multiplex adenine base editing in plants using new TadA variants. Mol Plant 14(5):722–731
Yang L, Machin F, Wang S, Saplaoura E, Kragler F (2023) Heritable transgene-free genome editing in plants by grafting of wild-type shoots to transgenic donor rootstocks. Nat Biotechnol. https://doi.org/10.1038/s41587-022-01585-8
Ye S, Enghiad B, Zhao H, Takano E (2020) Fine-tuning the regulation of Cas9 expression levels for efficient CRISPR-Cas9 mediated recombination in Streptomyces. J Ind Microbiol Biotechnol 47(4–5):413–423
Ye Q, Meng X, Chen H, Wu J, Zheng L, Shen C, Guo D, Zhao Y, Liu J, Xue Q (2022) Construction of genic male sterility system by CRISPR/Cas9 editing from model legume to alfalfa. Plant Biotechnol J 20(4):613
Yue E, Cao H, Liu B (2020) OsmiR535, a potential genetic editing target for drought and salinity stress tolerance in Oryza sativa. Plants 9(10):1337
Zeng Y, Wen J, Zhao W, Wang Q, Huang W (2020) Rational improvement of rice yield and cold tolerance by editing the three genes OsPIN5b, GS3, and OsMYB30 with the CRISPR–Cas9 system. Front Plant Sci 10:1663
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, Van Der Oost J, Regev A (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163(3):759–771
Zhan X, Zhang F, Zhong Z, Chen R, Wang Y, Chang L, Bock R, Nie B, Zhang J (2019) Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol J 17(9):1814–1822
Zhan X, Liu W, Nie B, Zhang F, Zhang J (2023) Cas13d-mediated multiplex RNA targeting confers a broad-spectrum resistance against RNA viruses in potato. Communications Biology 6(1):855. https://doi.org/10.1038/s42003-023-05205-2
Zhang C, Konermann S, Brideau NJ, Lotfy P, Wu X, Novick SJ, Strutzenberg T, Griffin PR, Hsu PD, Lyumkis D (2018a) Structural basis for the RNA-guided ribonuclease activity of CRISPR-Cas13d. Cell 175(1):212-223.e217
Zhang J, Zhang H, Botella JR, Zhu JK (2018b) Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J Integr Plant Biol 60(5):369–375. https://doi.org/10.1111/jipb.12620
Zhang T, Zheng Q, Yi X, An H, Zhao Y, Ma S, Zhou G (2018c) Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol J 16(8):1415–1423
Zhang A, Liu Y, Wang F, Li T, Chen Z, Kong D, Bi J, Zhang F, Luo X, Wang J, Tang J, Yu X, Liu G, Luo L (2019) Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol Breeding 39(3):47. https://doi.org/10.1007/s11032-019-0954-y
Zhang H, Li T, Sun Y, Yang H (2021a) Perfecting targeting in CRISPR. Annu Rev Genet 55:453–477
Zhang S, Zhang R, Gao J, Song G, Li J, Li W, Qi Y, Li Y, Li G (2021b) CRISPR/Cas9-mediated genome editing for wheat grain quality improvement. Plant Biotechnol J 19(9):1684
Zhou Y, Xu S, Jiang N, Zhao X, Bai Z, Liu J, Yao W, Tang Q, Xiao G, Lv C (2022) Engineering of rice varieties with enhanced resistances to both blast and bacterial blight diseases via CRISPR/Cas9. Plant Biotechnol J 20(5):876–885
Acknowledgements
We acknowledge the funders of this research and the laboratory members of CBE at NJFU.
Funding
This research work was funded by the National Natural Science Foundation of China (No. 31971682) and the Research Startup Fund for High-Level and Highly-Educated Talents of Nanjing Forestry University.
Author information
Authors and Affiliations
Contributions
DH, KY, and YL conceived and designed the idea. DH wrote the manuscript. YR, KY, JC and YL revised and edited the manuscript. YL sourced funding. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hwarari, D., Radani, Y., Ke, Y. et al. CRISPR/Cas genome editing in plants: mechanisms, applications, and overcoming bottlenecks. Funct Integr Genomics 24, 50 (2024). https://doi.org/10.1007/s10142-024-01314-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-024-01314-1