Abstract
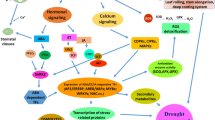
Development of abiotic stress tolerant rice cultivars is necessary for sustainable rice production under the scenario of global climate change, dwindling fresh water resources and increase in salt affected areas. Several genes from rice have been functionally validated by using EMS mutants and transgenics. Often, many of these desirable alleles are not available indica rice which is mainly cultivated, and where available, introgression of these alleles into elite cultivars is a time and labour intensive process, in addition to the potential introgression of non-desirable genes due to linkage. CRISPR-Cas technology helps development of elite cultivars with desirable alleles by precision gene editing. Hence, this study was carried out to create mutant alleles of drought and salt tolerance (DST) gene by using CRISPR-Cas9 gene editing in indica rice cv. MTU1010. We used two different gRNAs to target regions of DST protein that might be involved in protein–protein interaction and successfully generated different mutant alleles of DST gene. We selected homozygous dst mutant with 366 bp deletion between the two gRNAs for phenotypic analysis. This 366 bp deletion led to the deletion of amino acid residues from 184 to 305 in frame, and hence the mutant was named as dst∆184–305. The dst∆184–305 mutation induced by CRISPR-Cas9 method in DST gene in indica rice cv. MTU1010 phenocopied EMS-induced dst (N69D) mutation reported earlier in japonica cultivar. The dst∆184–305 mutant produced leaves with broader width and reduced stomatal density, and thus enhanced leaf water retention under dehydration stress. Our study showed that the reduction in stomatal density in loss of function mutants of dst is, at least, in part due to downregulation of stomatal developmental genes SPCH1, MUTE and ICE1. The Cas9-free dst∆184–305 mutant exhibited moderate level tolerance to osmotic stress and high level of salt stress in seedling stage. Thus, dst mutant alleles generated in this study will be useful for improving drought and salt tolerance and grain yield in indica rice cultivars.






Similar content being viewed by others
References
Birla DS, Malik K, Sainger M, Chaudhary D, Jaiwal R, Jaiwal PK (2017) Progress and challenges in improving the nutritional quality of rice (Oryza sativa L.). Crit Rev Food Sci Nutr 57:2455–2481
Chen K, Wang Y, Zhang R, Zhang H, Gao C (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 70:667–697
Chu CC, Hill RD, Brule-Babel AI (1990) High frequency of pollen embryoid formation and plant regeneration in Triticum aestivum L. on monosaccharide containing media. Plant Sci 66:255–262
Cui LG, Shan JX, Shi M, Gao JP, Lin HX (2015) DCA1 acts as a transcriptional co-activator of DST and contributes to drought and salt tolerance in rice. PLoS Genet 11(10):e1005617. https://doi.org/10.1371/journal.pgen.1005617
Dehairs J, Talebi A, Cherifi Y, Swinnen JV (2016) CRISP-ID: decoding CRISPR mediated indels by Sanger sequencing. Sci Rep 6:28973. https://doi.org/10.1038/srep28973
Feng Z, Zhang B, Ding W, Liu X, Yang D, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu JK (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23:1229–1232
Hiei Y, Ishida Y, Komari T (2015) Rice, Indica (Oryza sativa L.). In: Wang K. (eds). Agrobacterium Protocols. Methods Mol Biol 1223. Springer, New York, NY
Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2004) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34(5):733–739
Hiscox JD, Israelstam GF (1979) A method for extraction of chlorophyll from leaf tissue without maceration. Can J Bot 57:1332–1334
Hua K, Tao X, Zhu JK (2019) Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol J 17:499–504
Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23:1805–1817
Jain RK, Khehra GS, Lee S-H, Blackhall NW, Marchant R, Davey MR, Power JB, Cocking EC, Gosal SS (1995) An improved procedure for plant regeneration from indica and japonica rice protoplasts. Plant Cell Rep 14:515–519
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651
Jun R, Xixun H, Keijan W, Chun W (2019) Development and application of CRISPR/Cas system in rice. Rice Sci 26(2):69–76
Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU (2008) SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20(7):1775–1785
Kim J (2018) Precision genome engineering through adenine and cytosine base editing. Nat Plants 4:148–151
Kumar KK, Maruthasalam S, Loganathan M, Sudhakar D, Balasubramanian P (2005) An improved Agrobacterium mediated transformation protocol for recalcitrant elite indica rice cultivars. Plant Mol Biol Rep 23:67–73
Kusumi K, Hirotsuka S, Kumamaru T, Iba K (2012) Increased leaf photosynthesis caused by elevated stomatal conductance in a rice mutant deficient in SLAC1, a guard cell anion channel protein. J Exp Bot 63:5635–5644
Li T, Liu B, Spalding MH, Weeks DP, Yang B (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30:390–392
Li S, Zhao B, Yuan D, Duan M, Qian Q, Tang L, Wang B, Liu X, Zhang J, Wang J, Sun J, Liu Z, Feng YQ, Yuan L, Li C (2013) Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc Natl Acad Sci USA 110:3167–3172
Li M, Li X, Zhou Z, Wu P, Fang M, Pan X, Lin Q, Luo W, Wu G, Li H (2016) Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci 7:377. https://doi.org/10.3389/fpls.2016.00377
Li Y, Xiao J, Chen L, Huang X, Cheng Z, Han B, Zhang Q, Wu C (2018) Rice functional genomics research: past decade and future. Mol Plant 11:359–380
Liu W, Xie X, Ma X, Li J, Chen J, Liu YG (2015) DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol Plant 8:1431–1433
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods San Diego Calif 25:402–408
Ma X, Chen L, Zhu Q, Chen Y, Liu YG (2015) Rapid decoding of sequence-specific nuclease-induced heterozygous and biallelic mutations by direct sequencing of PCR products. Mol Plant 8:1285–1287
Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK (2013) Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant 6:2008–2011
Meng X, Yu H, Zhang Y, Zhuang F, Song X, Gao S, Gao C, Li J (2017) Construction of a genome-wide mutant library in rice using CRISPR/Cas9. Mol Plant 10:1238–1241
Miao C, Xiao L, Hua K, Zou C, Zhao Y, Bressan RA, Zhu JK (2018) Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc Natl Acad Sci USA 115:6058–6063
Mishra R, Joshi RK, Zhao K (2018) Genome editing in rice: recent advances, challenges, and future implications. Front Plant Sci 9:1361. https://doi.org/10.3389/fpls.2018.01361
Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E, García-Casado G, Witters E, Inzé D, Long JA, De Jaeger G, Solano R, Goossens A (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464(7289):788–791
Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU (2007) Termination of asymmetric cell division and differentiation of stomata. Nature 445(7127):501–505
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8(6):e66428. https://doi.org/10.1371/journal.pone.0066428
Sahoo RK, Tuteja N (2012) Development of agrobacterium-mediated transformation technology for mature seed-derived callus tissues of indica rice cultivar IR64. GM Crops Food 3:123–128
Sahoo KK, Tripathi AK, Pareek A, Sopory SK, Singla-Pareek SL (2011) An improved protocol for efficient transformation and regeneration of diverse indica rice cultivars. Plant Methods 7:49. https://doi.org/10.1186/1746-4811-7-49
Singh R, Singh Y, Xalaxo S, Verulkar S, Yadav N, Singh S, Singh N, Prasad KSN, Kondayya K, Rao PVR, Rani MG, Anuradha T, Suraynarayana Y, Sharma PC, Krishnamurthy SL, Sharma SK, Dwivedi JL, Singh AK, Singh PK, Nilanjay Singh NK, Kumar R, Chetia SK, Ahmad T, Rai M, Perraju P, Pande A, Singh DN, Mandal NP, Reddy JN, Singh ON, Katara JL, Marandi B, Swain P, Sarkar RK, Singh DP, Mohapatra T, Padmawathi G, Ram T, Kathiresan RM, Paramsivam K, Nadarajan S, Thirumeni S, Nagarajan M, Singh AK, Vikram P, Kumar A, Septiningshih E, Singh US, Ismail AM, Mackill D, Singh NK (2016) From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Sci 242:278–287
Strickland SG, Nichol JW, McCaU CM, Stuart DA (1987) Effect of carbohydrate source on alfalfa somatic embryogenesis. Plant Sci 48:113–121
Tan L, Li X, Liu F, Sun X, Li C, Zhu Z, Fu Y, Cai H, Wang X, Xie D, Sun C (2008) Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet 40(11):1360–1364
Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16(2):533–543
Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45:523–539
Wang F, Wang C, Liu P, Lei P, Hao W, Gao Y, Liu YG, Zhao K (2016) Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 11(4):e0154027. https://doi.org/10.1371/journal.pone.0154027
Wu Z, Chen L, Yu Q, Zhou W, Gou X, Li J, Hou S (2019) Multiple transcriptional factors control stomata development in rice. New Phytol 223(1):220–232
Xu R, Li H, Qin R, Wang L, Li L, Wei P, Yang J (2014) Gene targeting using the Agrobacterium tumefaciens mediated CRISPR-Cas system in rice. Rice 7(1):5. https://doi.org/10.1186/s12284-014-0005-6
Xu R, Qin R, Li H, Li D, Li L, Wei P, Yang J (2017) Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol J 15:713–717
Yao W, Li G, Yu Y, Ouyang Y (2018) funRiceGenes dataset for comprehensive understanding and application of rice functional genes. Gigascience 7(1):1–9
Zhou H, Liu B, Weeks DP, Spalding MH, Yang B (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acid Res 42:10903–10914
Zoulias N, Harrison EL, Casson SA, Gray JE (2018) Molecular control of stomatal development. Biochem J 475(2):441–454
Acknowledgements
This work was supported by Indian Council of Agricultural Research (ICAR) -National Agricultural Science Fund, New Delhi, Grant No. NASF/CRISPR-Cas-7003/2018-19, and ICAR-Indian Agricultural Research Institute, New Delhi, Grant No. CRSCIARISIL20144047279.
Author information
Authors and Affiliations
Contributions
SKVV prepared gene constructs, optimized callus induction, transformation and developed GEd plants; PY and AW developed GE plants. RKV and SKV did molecular analysis of transgenic plants. SKVV and MVR wrote the manuscript. VC designed the experiments, helped in data analysis and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Santosh Kumar, V.V., Verma, R.K., Yadav, S.K. et al. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol Mol Biol Plants 26, 1099–1110 (2020). https://doi.org/10.1007/s12298-020-00819-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-020-00819-w