Abstract
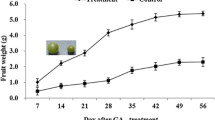
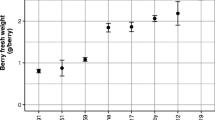
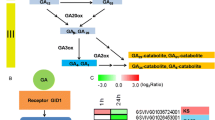
In grapes (Vitis vinifera L.), exogenous gibberellic acid (GA3) is applied at different stages of bunch development to achieve desirable bunch shape and berry size in seedless grapes used for table purpose. RNA sequence-based transcriptome analysis was used to understand the mechanism of GA3 action at cluster emergence, full bloom, and berry stage in table grape variety Thompson Seedless. At cluster emergence, rachis samples were collected at 6 and 24 h after application of GA3, whereas flower clusters and berry samples were collected at 6, 24, and 48 h after application at full bloom and 3–4 mm berry stages. Seven hundred thirty-three genes were differentially expressed in GA3-treated samples. At rachis and flower cluster stage respectively, 126 and 264 genes were found to be significantly differentially expressed within 6 h of GA3 application. The number of DEG reduced considerably at 24 h. However, at berry stage, major changes occurred even at 24 h and a number of DEGs at 6 and 24 h were 174 and 191, respectively. As compared to upregulated genes, larger numbers of genes were downregulated. Stage-specific response to the GA3 application was observed as evident from the unique set of DEGs at each stage and only a few common genes among three stages. Among the DEGs, 67 were transcription factors. Functional categorization and enrichment analysis revealed that several transcripts involved in sucrose and hexose metabolism, hormone and secondary metabolism, and abiotic and biotic stimuli were enriched in response to application of GA3. A high correlation was recorded for real-time PCR and transcriptome data for selected DEGs, thus indicating the robustness of transcriptome data obtained in this study for understanding the GA3 response at different stages of berry development in grape. Chromosomal localization of DEGs and identification of polymorphic microsatellite markers in selected genes have potential for their use in breeding for varieties with improved bunch architecture.










Similar content being viewed by others
References
Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311:91–94. https://doi.org/10.1126/science.1118642
Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, Lapierre C, Jouanin L (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23:1124–1137. https://doi.org/10.1105/tpc.110.082792
Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ (2002) Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 14:3073–3088. https://doi.org/10.1105/tpc.007773
Burdiak P, Rusaczonek A, Witoń D, Głów D, Karpiński S (2015) Cysteine-rich receptor-like kinase CRK5 as a regulator of growth, development, and ultraviolet radiation responses in Arabidopsis thaliana. J Exp Bot 66:3325–3337. https://doi.org/10.1093/jxb/erv143
Chai L, Li Y, Chen S, Perl A, Zhao F, Ma H (2014) RNA sequencing reveals high resolution expression change of major plant hormone pathway genes after young seedless grape berries treated with gibberellin. Plant Sci 229:215–224. https://doi.org/10.1016/j.plantsci.2014.09.010
Chen L-Q, Hou B-H, Lalonde S, Takanaga H, Hartung ML, Qu X-Q, Guo W-J, Kim J-G, Underwood W, Chaudhuri B, Chermak D, Antony G, White FF, Somerville SC, Mudgett MB, Frommer WB (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468:527–532 http://www.nature.com/nature/journal/v468/n7323/abs/nature09606.html#supplementary-information
Chen X, Lu S, Wang Y, Zhang X, Lv B, Luo L, Xi D, Shen J, Ma H, Ming F (2015) OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. Plant J 82:302–314. https://doi.org/10.1111/tpj.12819
Cheng C, Jiao C, Singer SD, Gao M, Xu X, Zhou Y, Li Z, Fei Z, Wang Y, Wang X (2015) Gibberellin-induced changes in the transcriptome of grapevine (Vitis labrusca × V. vinifera) cv. Kyoho flowers. BMC Genomics 16:128. https://doi.org/10.1186/s12864-015-1324-8
Choi Y-J, Hur YY, Jung S-M, Kim S-H, Noh J-H, Park S-J, Park K-S, Yun H-K (2013) Transcriptional analysis of Dehydrin1 genes responsive to dehydrating stress in grapevines. Hortic Environ Biotechnol 54:272–279. https://doi.org/10.1007/s13580-013-0094-y
Chundawat BS, Singh R (1980) Effect of growth regulators on phalsa (Grewia asiatica L.) I. Growth and fruiting. Indian J Hortic 37:124–131
Correa J, Mamani M, Munoz-Espinoza C, Laborie D, Munoz C, Pinto M, Hinrichsen P (2014) Heritability and identification of QTLs and underlying candidate genes associated with the architecture of the grapevine cluster (Vitis vinifera L.) Theor Appl Genet 127:1143–1162. https://doi.org/10.1007/s00122-014-2286-y
Correa J, Ravest G, Laborie D, Mamani M, Torres E, Muñoz C, Pinto M, Hinrichsen P (2015) Quantitative trait loci for the response to gibberellic acid of berry size and seed mass in tablegrape (Vitis vinifera L.) Aust J Grape Wine Res 21:496–507. https://doi.org/10.1111/ajgw.12141
De Smet I, Signora L, Beeckman T, Inzé D, Foyer CH, Zhang H (2003) An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J 33:543–555. https://doi.org/10.1046/j.1365-313X.2003.01652.x
Derbyshire P, McCann MC, Roberts K (2007) Restricted cell elongation in Arabidopsis hypocotyls is associated with a reduced average pectin esterification level. BMC Plant Biol 7:31. https://doi.org/10.1186/1471-2229-7-31
Dokoozlian NK, Peacock WL (2001) Gibberellic acid applied at bloom reduces fruit set and improves size of ‘Crimson Seedless’ table grapes. Hortscience 36:706–709
Espinoza A, Contreras D, Orellana M, Pérez R, Aguirre C, Castro A, Riquelme A, Fichet T, Pinto M, Hinrichsen P (2009) Modulation by gibberellic acid of aquaporin genes expression during berry development of grapevine (Vitis vinifera L.) Acta Hortic 827:355–362
Estornell LH, Agustí J, Merelo P, Talón M, Tadeo FR (2013) Elucidating mechanisms underlying organ abscission. Plant Sci 199-200:48–60. https://doi.org/10.1016/j.plantsci.2012.10.008
Farrokhi N, Burton RA, Brownfield L, Hrmova M, Wilson SM, Bacic A, Fincher GB (2006) Plant cell wall biosynthesis: genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol J 4:145–167. https://doi.org/10.1111/j.1467-7652.2005.00169.x
Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59:387–415. https://doi.org/10.1146/annurev.arplant.59.032607.092740
Fu X, Harberd NP (2003) Auxin promotes arabidopsis root growth by modulating gibberellin response. Nature 421:740–743
Fukazawa J, Nakata M, Ito T, Yamaguchi S, Takahashi Y (2010) The transcription factor RSG regulates negative feedback of NtGA20ox1 encoding GA 20-oxidase. Plant J 62:1035–1045. https://doi.org/10.1111/j.1365-313X.2010.04215.x
Grienenberger E, Besseau S, Geoffroy P, Debayle D, Heintz D, Lapierre C, Pollet B, Heitz T, Legrand M (2009) A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J 58:246–259. https://doi.org/10.1111/j.1365-313X.2008.03773.x
Heinrich M, Hettenhausen C, Lange T, Wünsche H, Fang J, Baldwin IT, Wu J (2013) High levels of jasmonic acid antagonize the biosynthesis of gibberellins and inhibit the growth of Nicotiana attenuata stems. Plant J 73:591–606. https://doi.org/10.1111/tpj.12058
Jan A, Komatsu S (2006) Functional characterization of gibberellin-regulated genes in rice using microarray system. Genomics Proteomics Bioinformatics 4:137–144. https://doi.org/10.1016/S1672-0229(06)60026-0
Kasimatis AN, Swanson FH, Vilas E, Peacock WL, Leavitt GM (1979) The relation of bloom-applied gibberellic acid to the yield and quality of Thompson Seedless raisins. Am J Enol Vitic 30:224–226
Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23:1647–1656
Lashbrooke JG, Young PR, Dockrall SJ, Vasanth K, Vivier MA (2013) Functional characterisation of three members of the Vitis vinifera L. carotenoid cleavage dioxygenase gene family. BMC Plant Biol 13:1–17. https://doi.org/10.1186/1471-2229-13-156
Lee C, Teng Q, Zhong R, Ye Z-H (2012) Arabidopsis GUX proteins are glucuronyltransferases responsible for the addition of glucuronic acid side chains onto xylan. Plant Cell Physiol 53:1204–1216. https://doi.org/10.1093/pcp/pcs064
Li R, Wang W, Wang W, Li F, Wang Q, Xu Y, Wang S (2015) Overexpression of a cysteine proteinase inhibitor gene from Jatropha curcas confers enhanced tolerance to salinity stress. Electron J Biotechnol 18:368–375. https://doi.org/10.1016/j.ejbt.2015.08.002
Li J, Yu X, Lou Y, Wang L, Slovin JP, Xu W, Wang S, Zhang C (2015) Proteomic analysis of the effects of gibberellin on increased fruit sink strength in Asian pear (Pyrus pyrifolia). Sci Hortic 195:25–36. https://doi.org/10.1016/j.scienta.2015.08.035
Liu X, Baird WV (2003) The ribosomal small-subunit protein S28 gene from Helianthus annuus (Asteraceae) is down-regulated in response to drought, high salinity, and abscisic acid. Am J Bot 90:526–531. https://doi.org/10.3732/ajb.90.4.526
Looney NE, Wood DF (1977) Some cluster thinning and gibberellic acid effects on fruit set, berry size, vine growth and yield of de Chaunac grapes. Can J Plant Sci (Ottawa) 57:653–659
Lu K, Liang S, Wu Z, Bi C, Yu Y-T, Ma Y, Wang X-F, Zhang D-P (2016) Overexpression of an Arabidopsis cysteine-rich receptor-like protein kinase, CRK5, enhances abscisic acid sensitivity and confers drought tolerance. J Exp Bot 67:5009–5027. https://doi.org/10.1093/jxb/erw266
Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2004) dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J 37:720–729. https://doi.org/10.1111/j.1365-313X.2003.01998.x
Mao D, Yu F, Li J, Tan D, Li J, Liu Y, Li X, Dong M, Chen L, Li D (2015) FERONIA receptor kinase interacts with S-adenosylmethionine synthetase and suppresses S-adenosylmethionine production and ethylene biosynthesis in Arabidopsis. Plant Cell Environ 38:2566–2574
May P (2000) From bud to berry, with special reference to inflorescence and bunch morphology in Vitis vinifera L. Aust J Grape Wine Res 6:82–98. https://doi.org/10.1111/j.1755-0238.2000.tb00166.x
Molitor D, Rothmeier M, Behr M, Fischer S, Hoffmann L, Evers D (2011) Crop cultural and chemical methods to control grey mould on grapes. Vitis 50
Molitor D, Behr M, Hoffmann L, Evers D (2012a) Benefits and drawbacks of pre-bloom applications of gibberellic acid (GA3) for stem elongation in Sauvignon Blanc. South Afr J Enol Vitic Stellenbosch 33:198–202
Molitor D, Behr M, Hoffmann L, Evers D (2012b) Impact of grape cluster division on cluster morphology and bunch rot epidemic. Am J Enol Vitic 63:508–514. https://doi.org/10.5344/ajev.2012.12041
Mosesian RM, Nelson KE (1968) Effect on ‘Thompson Seedless’ fruit of gibberellic acid bloom sprays and double girdling. Am J Enol Vitic 19:37–46
Nakata M, Yuasa T, Takahashi Y, Ishida S (2009) CDPK1, a calcium-dependent protein kinase, regulates transcriptional activator RSG in response to gibberellins. Plant Signal Behav 4:372–374
Oulkar DP, Banerjee K, Ghaste MS, Ramteke SD, Naik DG, Patil SB, Jadhav MR, Adsule PG (2011) Multiresidue analysis of multiclass plant growth regulators in grapes by liquid chromatography/tandem mass spectrometry. J AOAC Int 94:968–977
Pérez FJ, Gómez M (2000) Possible role of soluble invertase in the gibberellic acid berry-sizing effect in sultana grape. Plant Growth Regul 30:111–116. https://doi.org/10.1023/a:1006318306115
Petti C, Hirano K, Stork J, DeBolt S (2015) Mapping of a cellulose-deficient mutant named dwarf1-1 in sorghum bicolor to the green revolution gene gibberellin20-oxidase reveals a positive regulatory association between gibberellin and cellulose biosynthesis. Plant Physiol 169:705–716. https://doi.org/10.1104/pp.15.00928
Puig S (2014) Function and regulation of the plant COPT family of high-affinity copper transport proteins. Adv Bot 2014:9–9. https://doi.org/10.1155/2014/476917
Richter R, Behringer C, Müller IK, Schwechheimer C (2010) The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev 24:2093–2104. https://doi.org/10.1101/gad.594910
Robertson M (2004) Two transcription factors are negative regulators of gibberellin response in the HvSPY-signaling pathway in barley aleurone. Plant Physiol 136:2747–2761. https://doi.org/10.1104/pp.104.041665
Roper TR, Williams LE (1989) Net CO2 assimilation and carbohydrate partitioning of grapevine leaves in response to trunk girdling and gibberellic acid application. Plant Physiol 89:1136–1140
Schulz P, Herde M, Romeis T (2013) Calcium-dependent protein kinases: hubs in plant stress signaling and development. Plant Physiol 163:523–530. https://doi.org/10.1104/pp.113.222539
Shavrukov YN, Dry IB, Thomas MR (2004) Inflorescence and bunch architecture development in Vitis vinifera L. Aust J Grape Wine Res 10:116–124. https://doi.org/10.1111/j.1755-0238.2004.tb00014.x
Street IH, Shah PK, Smith AM, Avery N, Neff MM (2008) The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J 54:1–14. https://doi.org/10.1111/j.1365-313X.2007.03393.x
Tello J, Aguirrezábal R, Hernáiz S, Larreina B, Montemayor MI, Vaquero E, Ibáñez J (2015) Multicultivar and multivariate study of the natural variation for grapevine bunch compactness. Aust J Grape Wine Res 21:277–289. https://doi.org/10.1111/ajgw.12121
Tello J, Torres-Pérez R, Grimplet J, Ibáñez J (2016) Association analysis of grapevine bunch traits using a comprehensive approach. Theor Appl Genet 129:227–242. https://doi.org/10.1007/s00122-015-2623-9
Ugare B, Banerjee K, Ramteke SD, Pradhan S, Oulkar DP, Utture SC, Adsule PG (2013) Dissipation kinetics of forchlorfenuron, 6-benzyl aminopurine, gibberellic acid and ethephon residues in table grapes (Vitis vinifera). Food Chem 141:4208–4214. https://doi.org/10.1016/j.foodchem.2013.06.111
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115. https://doi.org/10.1093/nar/gks596
Upadhyay A, Aher LB, Shinde MP, Mundankar KY, Datre A, Karibasappa GS (2013) Microsatellite analysis to rationalize grape germplasm in India and development of a molecular database. Plant Genet Resour 11:225–233. https://doi.org/10.1017/S1479262113000117
Upadhyay A, Jogaiah S, Maske SR, Kadoo NY, Gupta VS (2015) Expression of stable reference genes and SPINDLY gene in response to gibberellic acid application at different stages of grapevine development. Biol Plant 59:436–444. https://doi.org/10.1007/s10535-015-0521-2
Wang Z, Zhao F, Zhao X, Ge H, Chai L, Chen S, Perl A, Ma H (2012) Proteomic analysis of berry-sizing effect of GA3 on seedless Vitis vinifera L. Proteomics 12:86–94. https://doi.org/10.1002/pmic.201000668
Weaver RJ (1958) Effect of gibberellic acid on fruit set and berry enlargement in seedless grapes of Vitis vinifera. Nature 181:851–852
Weaver RJ (1975) Effect of time of application of potassium gibberellate on cluster development of ‘Zinfandel’ grapes. Vitis 14:97–102
Weaver RJ, McCune S (1959) Effect of gibberellin on seedless Vitis vinifera. Hilgardia 29:247–275. https://doi.org/10.3733/hilg.v29n06p247
Woodger FJ, Millar A, Murray F, Jacobsen JV, Gubler F (2003) The role of GAMYB transcription factors in GA-regulated gene expression. J Plant Growth Regul 22:176–184. https://doi.org/10.1007/s00344-003-0025-8
Wu A-M, Lv S-Y, Liu J-Y (2007) Functional analysis of a cotton glucuronosyltransferase promoter in transgenic tobaccos. Cell Res 17:174–183
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–W322. https://doi.org/10.1093/nar/gkr483
Zavala JA, Casteel CL, Nabity PD, Berenbaum MR, DeLucia EH (2009) Role of cysteine proteinase inhibitors in preference of Japanese beetles (Popillia japonica) for soybean (Glycine max) leaves of different ages and grown under elevated CO2. Oecologia 161:35–41. https://doi.org/10.1007/s00442-009-1360-7
Zeng H, Xu L, Singh A, Wang H, Du L, Poovaiah BW (2015) Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front Plant Sci 6:600. https://doi.org/10.3389/fpls.2015.00600
Zhang S, Cai Z, Wang X (2009) The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc Natl Acad Sci 106:4543–4548. https://doi.org/10.1073/pnas.0900349106
Zhao J, Favero DS, Peng H, Neff MM (2013) Arabidopsis thaliana AHL family modulates hypocotyl growth redundantly by interacting with each other via the PPC/DUF296 domain. Proc Natl Acad Sci 110:E4688–E4697. https://doi.org/10.1073/pnas.1219277110
Acknowledgements
This work was funded by the Department of Biotechnology, Government of India, New Delhi, under the grant no. BT/PR4856/PBD/16/967/2012.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(XLSX 90 kb)
Rights and permissions
About this article
Cite this article
Upadhyay, A., Maske, S., Jogaiah, S. et al. GA3 application in grapes (Vitis vinifera L.) modulates different sets of genes at cluster emergence, full bloom, and berry stage as revealed by RNA sequence-based transcriptome analysis. Funct Integr Genomics 18, 439–455 (2018). https://doi.org/10.1007/s10142-018-0605-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-018-0605-0