Abstract
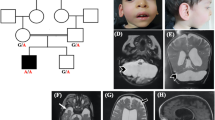
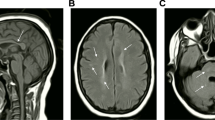
Pathogenic variants in L1CAM, the gene encoding the L1 cell adhesion molecule, are responsible for a wide clinical spectrum including X-linked hydrocephalus with stenosis of the Sylvius aqueduct, MASA syndrome (mental retardation, aphasia, shuffling gait, adducted thumbs), and a form of spastic paraplegia (SPG1). A moderate phenotype with mild intellectual disability (ID) and X-linked partial corpus callosum agenesis (CCA) has only been related to L1CAM in one family. We report here a second family, including 5 patients with mild to moderate ID and partial CCA without signs usually associated with L1CAM pathogenic variations (such as hydrocephalus, pyramidal syndrome, thumb adductus, aphasia). We identified a previously unreported c.3226A > C transversion leading to a p.Thr1076Pro amino acid substitution in the fifth fibronectin type III domain (FnIII) of the protein which co-segregates with the phenotype within the family. We performed in vitro assays to assess the pathogenic status of this variation. First, the expression of the novel p.Thr1076Pro mutant in COS7 cells resulted in endoplasmic reticulum (ER) retention and reduced L1CAM cell surface expression, which is expected to affect both L1CAM-mediated cell-cell adhesion and neurite growth. Second, immunoblotting techniques showed that the immature form of the L1CAM protein was increased, indicating that this variation led to a lack of maturation of the protein. ID associated with CCA is not a common clinical presentation of L1CAM pathogenic variants. Genome-wide analyses will identify such variations and it is important to acknowledge this atypical phenotype.



Similar content being viewed by others
Data availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References
Brown WS, Jeeves MA, Dietrich R, Burnison DS (1999) Bilateral field advantage and evoked potential interhemispheric transmission in commissurotomy and callosal agenesis. Neuropsychologia 37(10):1165–1180
Paul LK, Van Lancker-Sidtis D, Schieffer B, Dietrich R, Brown WS (2003) Communicative deficits in agenesis of the corpus callosum: nonliteral language and affective prosody. Brain Lang 85(2):313–324
des Portes V, Rolland A, Velazquez-Dominguez J, Peyric E, Cordier M-P, Gaucherand P et al (2018) Outcome of isolated agenesis of the corpus callosum: a population-based prospective study. Eur J Paediatr Neurol 22(1):82–92
Marsh APL, Heron D, Edwards TJ, Quartier A, Galea C, Nava C, Rastetter A, Moutard ML, Anderson V, Bitoun P, Bunt J, Faudet A, Garel C, Gillies G, Gobius I, Guegan J, Heide S, Keren B, Lesne F, Lukic V, Mandelstam SA, McGillivray G, McIlroy A, Méneret A, Mignot C, Morcom LR, Odent S, Paolino A, Pope K, Riant F, Robinson GA, Spencer-Smith M, Srour M, Stephenson SEM, Tankard R, Trouillard O, Welniarz Q, Wood A, Brice A, Rouleau G, Attié-Bitach T, Delatycki MB, Mandel JL, Amor DJ, Roze E, Piton A, Bahlo M, Billette de Villemeur T, Sherr EH, Leventer RJ, Richards LJ, Lockhart PJ, Depienne C (2017) Mutations in DCC cause isolated agenesis of the corpus callosum with incomplete penetrance. Nat Genet 49(4):511–514
Marsh APL, Edwards TJ, Galea C, Cooper HM, Engle EC, Jamuar SS, Méneret A, Moutard ML, Nava C, Rastetter A, Robinson G, Rouleau G, Roze E, Spencer-Smith M, Trouillard O, Billette de Villemeur T, Walsh CA, Yu TW, IRC5 Consortium, Heron D, Sherr EH, Richards LJ, Depienne C, Leventer RJ, Lockhart PJ (2018) DCC mutation update: congenital mirror movements, isolated agenesis of the corpus callosum, and developmental split brain syndrome. Hum Mutat 39(1):23–39
Adle-Biassette H, Saugier-Veber P, Fallet-Bianco C, Delezoide A-L, Razavi F, Drouot N et al (2013) Neuropathological review of 138 cases genetically tested for X-linked hydrocephalus: evidence for closely related clinical entities of unknown molecular bases. Acta Neuropathol (Berl) 126(3):427–442
Vos YJ, de Walle HEK, Bos KK, Stegeman JA, Ten Berge AM, Bruining M et al (2010) Genotype-phenotype correlations in L1 syndrome: a guide for genetic counselling and mutation analysis. J Med Genet. mars 47(3):169–175
Weller S, Gärtner J (2001) Genetic and clinical aspects of X-linked hydrocephalus (L1 disease): mutations in the L1CAM gene. Hum Mutat 18(1):1–12
Djabali M, Mattei MG, Nguyen C, Roux D, Demengeot J, Denizot F, Moos M, Schachner M, Goridis C, Jordan BR (1990) The gene encoding L1, a neural adhesion molecule of the immunoglobulin family, is located on the X chromosome in mouse and man. Genomics 7(4):587–593
Moos M, Tacke R, Scherer H, Teplow D, Früh K, Schachner M (1988) Neural adhesion molecule L1 as a member of the immunoglobulin superfamily with binding domains similar to fibronectin. Nature 334(6184):701–703
Williams EJ, Furness J, Walsh FS, Doherty P (1994) Activation of the FGF receptor underlies neurite outgrowth stimulated by L1, N-CAM, and N-cadherin. Neuron 13(3):583–594
Schmid RS, Maness PF (2008) L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Curr Opin Neurobiol 18(3):245–250
Barbin G, Aigrot MS, Charles P, Foucher A, Grumet M, Schachner M et al (2004) Axonal cell-adhesion molecule L1 in CNS myelination. Neuron Glia Biol 1(1):65–72
Wei CH, Ryu SE (2012) Homophilic interaction of the L1 family of cell adhesion molecules. Exp Mol Med. 44(7):413–423
Vos YJ, Hofstra RMW (2010) An updated and upgraded L1CAM mutation database. Hum Mutat 31(1):E1102–E1109
Basel-Vanagaite L, Straussberg R, Friez MJ, Inbar D, Korenreich L, Shohat M et al (2006) Expanding the phenotypic spectrum of L1CAM-associated disease. Clin Genet. mai 69(5):414–419
Redin C, Le Gras S, Mhamdi O, Geoffroy V, Stoetzel C, Vincent M-C et al (2012) Targeted high-throughput sequencing for diagnosis of genetically heterogeneous diseases: efficient mutation detection in Bardet-Biedl and Alström syndromes. J Med Genet 49(8):502–512
Geoffroy V, Pizot C, Redin C, Piton A, Vasli N, Stoetzel C et al (2015) VaRank: a simple and powerful tool for ranking genetic variants. Peer J 3:e796
Marx M, Diestel S, Bozon M, Keglowich L, Drouot N, Bouché E, Frebourg T, Minz M, Saugier-Veber P, Castellani V, Schäfer MKE (2012) Pathomechanistic characterization of two exonic L1CAM variants located in trans in an obligate carrier of X-linked hydrocephalus. Neurogenetics. févr 13(1):49–59
Rünker AE, Bartsch U, Nave K-A, Schachner M (2003) The C264Y Missense Mutation in the extracellular domain of L1 impairs protein trafficking in vitro and in vivo. J Neurosci 23(1):277–286
De Angelis E (2002) Disease-associated mutations in L1 CAM interfere with ligand interactions and cell-surface expression. Hum Mol Genet 11(1):1–12
De Angelis E (1999) Pathological missense mutations of neural cell adhesion molecule L1 affect homophilic and heterophilic binding activities. Embo J 18(17):4744–4753
Bateman A, Jouet M, MacFarlane J, Du JS, Kenwrick S, Chothia C (1996) Outline structure of the human L1 cell adhesion molecule and the sites where mutations cause neurological disorders. Embo J 15(22):6050–6059
Jouet M, Feldman E, Yates J, Donnai D, Paterson J, Siggers D, Kenwrick S (1993) Refining the genetic location of the gene for X linked hydrocephalus within Xq28. J Med Genet 30(3):214–217
Jouet M, Moncla A, Paterson J, McKeown C, Fryer A, Carpenter N et al (1995) New domains of neural cell-adhesion molecule L1 implicated in X-linked hydrocephalus and MASA syndrome. Am J Hum Genet 56(6):1304–1314
Richards S, Aziz N, Bale S, Bick D, Das S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–423
Gu SM, Orth U, Veske A, Enders H, Klunder K, Schlosser M et al (1996) Five novel mutations in the L1CAM gene in families with X linked hydrocephalus. J Med Genet 33(2):103–106
Acknowledgments
We are sincerely grateful to the family members for their participation in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare. Several authors of this publication (RT, APi, BG, DS, PE, PSV, APu) are members of the European Reference Network for Developmental Anomalies and Intellectual Disability (ERN-ITHACA).
Ethics approval
The authors performed the study in accordance to local guidance.
Consent to participate
Informed consent was obtained from patients and parents.
Consent for publication
Patients and parents signed informed consent regarding publishing the clinical data.
Code availability
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Figure 1
Confocal images of COS7 cells transfected with L1-WT, p.Trp635Cys, p.Thr1076Pro, and p.Thr1076Ser mutants. Double immunofluorescent labeling of L1 (red) and endoplasmic reticulum (ER) marker KDEL (green) were performed. The p.Trp635Cys mutant is a previously reported pathogenic mutant of L1CAM, serving as a positive control. The white asterisk depicts cells with yellow staining reflecting L1 accumulation in the ER, which is an increase in p.Trp635Cys and p.Thr1076Pro. Scale, 10 μm (PNG 3813 kb)
Rights and permissions
About this article
Cite this article
Bousquet, I., Bozon, M., Castellani, V. et al. X-linked partial corpus callosum agenesis with mild intellectual disability: identification of a novel L1CAM pathogenic variant. Neurogenetics 22, 43–51 (2021). https://doi.org/10.1007/s10048-020-00629-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-020-00629-y