Abstract
Background
CCDC88C is a ubiquitously expressed protein with multiple functions, including roles in cell polarity and the development of dendrites in the nervous system. Bi-allelic mutations in the CCDC88C gene cause autosomal recessive congenital hydrocephalus (OMIM #236600). Studies recently linked heterozygous mutations in CCDC88C to the development of the late-onset spinocerebellar ataxia type 40 (OMIM #616053).
Case presentation
A 48-year-old Sudanese female presented with pure early onset hereditary spastic paraplegia. Exome sequencing, in-silico analysis, and Sanger sequencing identified the heterozygous NM_001080414.4:c.1993G > A (p.E665K) variant in CCDC88C as a potential cause of her illness. To explore the pathogenicity of the NM_001080414.4:c.1993G > A (p.E665K) variant, we expressed it in human embryonic kidney 293 cells and assessed its effects on apoptosis. In our experiment, NM_001080414.4:c.1993G > A (p.E665K) induced JNK hyper-phosphorylation and enhanced apoptosis. In contrast to previous reports, our patient developed neurological symptoms in early childhood and showed neither features of cerebellar ataxia, extrapyramidal signs, nor evidence of intellectual involvement.
Conclusion
We, herein, heighlighted the possibility of extending the phenotype associated with variants in CCDC88C to include early-onset pure hereditary spastic paraplegia.
Similar content being viewed by others
Background
Hereditary spastic paraplegia and hereditary ataxia are distinct entities that share common clinical features and pathological mechanisms [1, 2]. Bi-allelic mutations in the CCDC88C gene are known to cause autosomal recessive non-syndromic congenital hydrocephalus (OMIM # 236600) [3]. Furthermore, two reports recently linked heterozygous mutations in CCDC88C to the development of the autosomal dominant spinocerebellar ataxia type 40 (SCA40; OMIM # 616053) [4, 5]. In the first report, the variant NM_001080414.4:c.1391G > A (p.R464H) caused a phenotype of ataxia, dysarthria, and pyramidal features, brisk reflexes in one patient and spastic lower limbs in a second patient, in a family from Hong Kong [4]. In the second report, the variant NM_001080414.4:c.127G > A (p.D43N) caused a phenotype of ataxia with parkinsonian features and dementia in a Polish family [5]. The age at onset in the patients from both families, when provided, was above 30 years [4, 5]. The severity of the ataxia ranged from severe in the Chinese family to relatively mild in the Polish family. However, unlike in the Chinese patients, the phenotype in the Polish family involved extrapyramidal features [4, 5].
We, herein, report for the first time a case of pure childhood-onset hereditary spastic paraplegia caused by the heterozygous variant NM_001080414:c.1993G > A (p.E665K) in the CCDC88C gene.
Case presentation
A 48-year-old Sudanese lady, coded as F83–581, presented with an abnormal gait as a manifestation of pure hereditary spastic paraplegia. Her condition started in early childhood with tip-toeing that progressed gradually in severity. At the age of 30 years, she could walk only using two sticks. She did not complain of any additional symptoms apart from occasional muscle cramps. Her parents were distantly related and had no family history of similar conditions. She was not on treatment. On examination, her lower limbs were spastic with severe weakness (power grade 3). There were bilateral deformities in the feet (pes equinovarus on the right and hammertoe on the left) and up-going plantar responses. Her upper limbs were normal except for mild spasticity and hyperreflexia on the right side. The patient (F83–581) had neither signs of cerebellar involvement nor evidence of sensory deficit. She was cooperative, oriented, and had no evidence of intellectual alteration. She could barely walk supported by two sticks, and her gait was spastic. Nerve conduction studies were normal. Brain magnetic resonance imaging (MRI) showed periventricular leukomalacia with scattered ischemic foci in the white matter, cerebellum, and right side of the pons. The isthmus of the corpus callosum was thin, but it could be a normal variant. We noted neither cerebral, brain stem, nor cerebellar atrophy, nor acute ischemic changes on the brain MRI (Fig. 1).
Brain MRI of the patient F83–581. a Sagittal T1 section showing thinning in the isthmus of the corpus callosum (arrow). b and c Axial T1 section showing periventricular white matter changes and scattered hyperintensities in the white matter, cerebellum, and right side of the pons (arrows). The cerebrum, brain stem, and cerebellum were not atrophied
We extracted DNA from the patient and four of her family members and investigated the patient and one of her healthy siblings, coded F83–582, using whole-exome sequencing [6]. Whole-exome sequencing of the patient revealed a heterozygous variant, NM_001080414.4:c.1993G > A (p.E665K) (rs956104232), in the CCDC88C gene that results in substituting Glutamate at position 665 of the protein for Lysine. Sift [7], Polyphen2 HDIV [8], Mutation Taster [9], Provean [10] and M-cap [11] embedded in VarAFT software [12] predicted this substitution as pathogenic with prediction scores of 0.002, 0.982, 1, − 3.21 and 0.069, respectively. Glutamate at position 665 of CCDC88C is highly conserved during evolution. The CADD score of 25 was also in favor of a pathogenic role of this change. We did not detect other convincing variants that could explain the phenotype in our patient. The variant NM_001080414.4:c.1993G > A (p.E665K) was reported once in the gnomAD v2.1.1 database in an individual of African ancestry and had a global allele frequency of 0.0000032 [13]. Using Sanger sequencing, we validated that the variant NM_001080414.4:c.1993G > A (p.E665K) was heterozygous in the patient and absent in her healthy family members (Fig. 2-I).
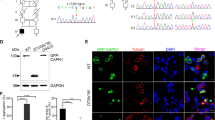
Segregation analysis, schematic representation of CCDC88C protein, and functional validation of the variant NM_001080414.4:c.1993G > A (p.E665K). I. The family pedigree and chromatogram showing the mutant variant NM_001080414.4:c.1993G > A (p.E665K) (filled arrowhead) heterozygous in the patient F83–581 (filled circle) and the wild-type variant at the same position (empty arrowheads) homozygous in the healthy controls (coded circles and square). M: mutant allele; WT: wild type allele. II. Schematic representation of CCDC88C protein based on the Uniprot entry Q9P219 showing the locations of the reported pathogenic heterozygous variants in the protein. CC: coiled-coil region; CH: calponin-homology domain. III. Expression of CCDC88CE665K mutant protein activated c-Jun N-terminal kinase (JNK) / caspase-3 apoptotic signaling pathway. a The wild-type and mutant pcDNA3.1(+)-myc-CCDC88C expression constructs were used to transfect Human Embryonic Kidney 293 (HEK293) cells to assess the effect of CCDC88CE665K mutant protein on the activity of JNK / caspase-3 signaling pathway. Similar to the other two CCDC88C mutant proteins (CCDC88CD43N and CCDC88CR464H) that were previously reported to activate JNK / caspase-3 signaling [4, 5], overexpression of CCDC88CE665K mutant protein caused a significant increase of JNK phosphorylation and caspase-3 cleavage. n = 3 biological replicates on 3 independent preparation of protein samples. Only representative blots were shown. b-d Quantification of the relative expression of p-JNK (b), cleaved caspase-3 (c), and CCDC88C (d) proteins. Error bars represent S.E.M. Statistical analysis was performed using one-way ANOVA followed by post hoc Tukey’s test. ns denotes no significant difference, * denotes p < 0.05, ** denotes p < 0.01 and *** denotes p < 0.001
To validate the pathogenicity of the NM_001080414.4:c.1993G > A (p.E665K) variant, we expressed the CCDC88C cDNA in human embryonic kidney (HEK) 293 cells and assessed its effect on c-Jun N-terminal kinase (JNK) / caspase-3 signaling pathway according to the presence or absence of the variant. Overexpressing CCDC88CE665K mutant protein caused a significant increase of JNK hyperphosphorylation and caspase-3 cleavage compared to the wild type protein, a pattern also seen when overexpressing the known SCA40 pathogenic proteins CCDC88CD43N and CCDC88CR464H (Fig. 2-III and supplementary data [6]). NM_001080414.4:c.1993G > A (p.E665K) was likely a de novo variant, though we did not have DNA samples from the parents. It had a low frequency in gnomAD database, predicted as pathogenic by multiple computational tools, and its pathogenicity was corroborated by functional studies, thus, fulfilling the criteria of likely pathogenic variants according to the American college of medical genetics and genomics guidelines for interpreting sequence variants published in 2015 [14]. We have submitted the variant to the Clinvar database (accession VCV000978819.2).
Discussion and conclusion
CCDC88C is a cytoplasmic protein ubiquitously expressed in mammals and has a variety of functions [15]. Up-to-date, mutations in CCDC88C are linked to monogenic neurological phenotypes by one of two mechanisms depending on the mutant allele, activation of hetero-trimeric G-proteins [16] and activation of JNK cascade [4].
Bi-allelic loss of function mutations in CCDC88C cause autosomal recessive congenital hydrocephalus (OMIM #236600) by disrupting the morphogenesis of the nervous system [3, 16]. On the other hand, gain-of-function mutations in CCDC88C cause SCA40 through mechanisms that induce JNK hyper-phosphorylation and prompt apoptosis [4].
In this report, we showed that a mutation in the CCDC88C gene induces JNK hyper-phosphorylation and the resulting activation of apoptosis in early-onset pure hereditary spastic paraplegia. Recently the borders between hereditary ataxia and hereditary spastic paraplegia have been questioned [2]. SCA40 is an emerging clinical entity identified up-to-date in only two families worldwide (Table 1), the first family was from China and the second family was from Poland [4, 5]. Cerebellar features were present in both families, though with different degrees of severity. Extrapyramidal features and dementia dominated the phenotype in the Polish family. Pyramidal involvement, hyperreflexia in one patient and spasticity in a second patient, was only evident in examining the Chinese family’s patients. The pathogenic mutation in the Polish family, NM_001080414.4:c.127G > A (p.D43N), is located in the calponin-homology domain of the CCDC88C protein. The mutations reported in our patient and in the Chinese patients, NM_001080414.4:c.1993G > A (p.E665K) and NM_001080414.4:c.1391G > A (p.R464H), respectively, are located in a coiled-coil domain in the protein (Fig. 2-II).
Although we did not have DNA samples from the patient’s parents, we assumed NM_001080414.4:c.1993G > A (p.E665K) was a de novo variant because the parents neither manifested the disease throughout their lives nor had a positive family history. However, we could not completely rebut the possibility of incomplete penetrance in the parents. The early age at onset in our patient compared to the two previously reported SCA40 families, adds another layer of complexity and suggests the presence of unknown modifiers. More studies are needed to delineate the phenotypes’ spectrum associated with CCDC88C mutations and investigate genotype-phenotype correlations.
In conclusion, this report suggests the extension of CCDC88C-associated phenotype to include early-onset pure hereditary spastic paraplegia.
Availability of data and materials
Supplementary data for this study are openly available in Figshare repository at https://doi.org/10.6084/m9.figshare.13020581.v1 [6].
Abbreviations
- SCA40:
-
Spinocerebellar ataxia type 40
- OMIM:
-
Online inheritance in man
- MRI:
-
Magnetic resonance imaging
- JNK:
-
c-Jun N-terminal kinase
- HEK:
-
Human Embryonic Kidney
References
Synofzik M, Schüle R. Overcoming the divide between ataxias and spastic paraplegias: Shared phenotypes, genes, and pathways. Mov Disord. 2017;32(3):332–45.
Parodi L, Coarelli G, Stevanin G, Brice A, Durr A. Hereditary ataxias and paraparesias: clinical and genetic update. Curr Opin Neurol. 2018;31(4):462–71.
Ekici AB, Hilfinger D, Jatzwauk M, Thiel CT, Wenzel D, Lorenz I, et al. Disturbed Wnt Signalling due to a mutation in CCDC88C causes an autosomal recessive non-Syndromic hydrocephalus with medial diverticulum. Mol Syndromol. 2010;1(3):99–112.
Tsoi H, Yu ACS, Chen ZS, Ng NKN, Chan AYY, Yuen LYP, et al. A novel missense mutation in CCDC88C activates the JNK pathway and causes a dominant form of spinocerebellar ataxia. J Med Genet. 2014;51(9):590–5.
Leńska-Mieciek M, Charzewska A, Królicki L, Hoffman-Zacharska D, Chen ZS, Lau KF, et al. Familial ataxia, tremor, and dementia in a polish family with a novel mutation in the CCDC88C gene. Mov Disord. 2019;34(1):142–4.
Ashraf Y, Chen ZS, E. Ahmed A, Emad S, Adil R, Abubaker R, et al. Supplementary file - a heterozygous mutation in CCDC88C gene causes early onset pure hereditary spastic paraplegia; 2020.
Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–4.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Vol. 7, Nature Methods; 2010. p. 248–9. NIH Public Access
Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575–6.
Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31(16):2745–7.
Jagadeesh KA, Wenger AM, Berger MJ, Guturu H, Stenson PD, Cooper DN, et al. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat Genet. 2016;48(12):1581–6.
Desvignes J-P, Bartoli M, Delague V, Krahn M, Miltgen M, Béroud C, et al. VarAFT: a variant annotation and filtration system for human next generation sequencing data. Nucleic Acids Res. 2018;46(W1):W545–53.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–43.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24.
Enomoto A, Ping J, Takahashi M. Girdin, a novel actin-binding protein, and its family of proteins possess versatile functions in the Akt and Wnt signaling pathways. Ann N Y Acad Sci. 2006;1086:169–84.
Marivin A, Morozova V, Walawalkar I, Leyme A, Kretov DA, Cifuentes D, et al. GPCR-independent activation of G proteins promotes apical cell constriction in vivo. J Cell Biol. 2019 May 1;218(5):1743–63.
Acknowledgments
The authors are grateful to the I-GenSeq ICM facility for exome processing.
Funding
This work was financially supported by the European Union grant (grant 779257 Solve-RD from the Horizon 2020 Research and Innovation Program) to Giovanni Stevanin and a CUHK Gerald Choa Neuroscience Centre grant (grant 7105306) to Ho Yin Edwin Chan. Mustafa A. Salih was supported by the Researchers Supporting Project (grant RSP-2020/38), King Saud University, Riyadh, Saudi Arabia. Zhefan Stephen Chen was supported by the Postdoctoral Fellowship in Clinical Neurosciences program between The Chinese University of Hong Kong and the University of Oxford (Nuffield Department of Clinical Neurosciences and Pembroke College) supported by the Lee Hysan Foundation. Ashraf Yahia’s Ph.D. was supported by the French embassy in Khartoum, Sudan, and the Ministry of Higher Education, Sudan. The funding bodies have neither role in the design, collection, analysis, and interpretation of data nor in writing the manuscript.
Author information
Authors and Affiliations
Contributions
AY, ZSC, LE, HYEC, and GS designed the study. AEA, AY, SOMAT, and MAS evaluated the patient and her investigations. AY, ZSC, SE, RA1, and RA2 collected the samples and relevant data and performed the experiments. AY, ZSC, HYEC, MAS, and GS interpreted the results. LE, AEA, HYEC, and GS supervised the study. AY and ZSC wrote the manuscript. All authors critically revised and approved the final version of the manuscript. All authors agreed to be accountable for all aspects of the work and ensured that questions related to the accuracy and the integrity of all parts of the work were appropriately investigated and resolved.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the ethics committee of the National University, Sudan (approval number NU-RECG200). All procedures in this study were performed following the ethical standards of the institutional research committee at the National University, Sudan, and with the 1975 Helsinki declaration and its later amendments. Informed written consent forms for participation were obtained from all participants.
Consent for publication
Signed informed consent forms were obtained from participants for publication of personal and medical details included in this report.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yahia, A., Chen, Z.S., Ahmed, A.E. et al. A heterozygous mutation in the CCDC88C gene likely causes early-onset pure hereditary spastic paraplegia: a case report. BMC Neurol 21, 78 (2021). https://doi.org/10.1186/s12883-021-02113-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12883-021-02113-y