Abstract
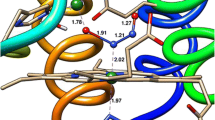
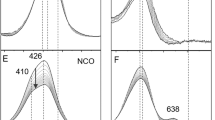
A re-investigation of the interaction with NO of the small tetraheme protein cytochrome c554 (C554) from Nitrosomonas europaea has shown that the 5-coordinate heme II of the two- or four-electron-reduced protein will nitrosylate reversibly. The process is first order in C554, first order in NO, and second-order overall. The rate constant for NO binding to the heme is 3000 ± 140 M−1s−1, while that for dissociation is 0.034 ± 0.009 s−1; the degree of protein reduction does not appear to significantly influence the nitrosylation rate. In contrast to a previous report (Upadhyay AK, et al. J Am Chem Soc 128:4330, 2006), this study found no evidence of C554-catalyzed NO reduction, either with \( {\text{C}}_{554}^{2 - } \) or with \( {\text{C}}_{554}^{4 - } . \) Some sub-stoichiometric oxidation of the lowest potential heme IV was detected when \( {\text{C}}_{554}^{4 - } \) was exposed to an excess of NO, but this is believed to arise from partial intramolecular electron transfer that generates {Fe(NO)}8 at heme II. The vacant heme II coordination site of C554 is crowded by three non-bonding hydrophobic amino acids. After replacing one of these (Phe156) with the smaller alanine, the nitrosylation rate for F156A2− and F156A4− was about 400× faster than for the wild type, though the rate of the reverse denitrosylation process was almost unchanged. Unlike in the wild-type C554, the 6-coordinate low-spin hemes of F156A4− oxidized over the course of several minutes after exposure to NO. Concomitant formation of N2O could explain this heme oxidation, though alternative explanations are equally plausible given the available data.

















Similar content being viewed by others
Abbreviations
- C554 :
-
Cytochrome c554
- NOR:
-
Nitric oxide reductase
- PaPy2Q:
-
N,N-bis(2-pyridylmethyl)-amine-N-ethyl-2-quinoline-2-carboxamide)
- DEANO:
-
1-(N,N-diethylamino)diazene-1-ium-1,2-diolate
- HEPES:
-
N-(2-hydroxyethyl) piperazine-N-ethanesulfonic acid
- SVD:
-
Singular value decomposition
References
Yamanaka T, Shinra M (1974) Cytochrome c-552 and cytochrome c-554 derived from Nitrosomonas europaea. J Biochem 75:1265–1273
Iverson TM, Arciero DM, Hsu BT, Logan MSP, Hooper AB, Rees DC (1998) Heme packing motifs revealed by the crystal structure of the tetra-heme cytochrome c554 from Nitrosomonas europaea. Nat Struct Biol 5(11):1005–1012
Iverson TM, Arciero DM, Hooper AB, Rees DC (2001) High-resolution structures of the oxidized and reduced states of cytochrome c554 from Nitrosomonas europaea. J Biol Inorg Chem 6(4):390–397
Upadhyay AK, Petasis DT, Arciero DM, Hooper AB, Hendrich MP (2003) Spectroscopic characterization and assignment of reduction potentials in the tetraheme cytochrome c(554) from Nitrosomonas europaea. J Am Chem Soc 125(7):1738–1747. https://doi.org/10.1021/Ja020922x
Whittaker M, Bergmann D, Arciero D, Hooper AB (2000) Electron transfer during the oxidation of ammonia by the chemolithotrophic bacterium Nitrosomonas europaea. Biochim Biophys Acta Bioenerg 1459(2–3):346–355
Kim HJ, Zatsman A, Upadhyay A, Whittaker M, Bergmann DJ, Hendrich MP, Hooper AB (2008) Membrane tetraheme cytochrome c m552 of the ammonia-oxidizing Nitrosomonas europaea: a ubiquinone reductase. Biochemistry 47:6539–6551
Upadhyay AK, Hooper AB, Hendrich MP (2006) NO reductase activity of the tetraheme cytochrome c(554) of Nitrosomonas europaea. J Am Chem Soc 128(13):4330–4337. https://doi.org/10.1021/Ja055183+
Hendriks J, Warne A, Gohlke U, Haltia T, Ludovici C, Lubben M, Saraste M (1998) The active site of bacterial nitric oxide reductase is a dinuclear iron center. Biochemistry 37:13102–13109
Wasser IM, de Vries S, Moenne-Loccoz P, Schroder I, Karlin KD (2002) Nitric oxide in biological denitrification: Fe/Cu metalloenzyme and metal complex NOx redox chemistry. Chem Rev 102:1201–1234
Hino T, Matsumoto Y, Nagano S, Sugimoto H, Fukumori Y, Murata T, Iwata S, Shiro Y (2010) Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science 230:1666–1670
Sato N, Ishii S, Sugimoto H, Hino T, Fukumori Y, Sako Y, Shiro Y, Tosha T (2014) Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes. Proteins 82:1258–1271
Shiro Y (2012) Structure and function of bacterial nitric oxide reductases. Biochim Biophys Acta 1817:1907–1913
Matsumura H, Chackraborty S, Reed J, Lu Y, Moenne-Loccoz P (2016) Effect of outer-sphere side chain substitutions on the fate of the trans-iron nitrosyl dimer in heme/nonheme engineered myoglobins (FeBMbs): insights into the mechanism of denitrifying NO reductases. Biochemistry 55:2091–2099
Lin Y-W, Yeung N, Gao Y-G, Miner KD, Tian S, Robinson H, Lu Y (2010) Roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin. PNAS 107:8581–8586
Abucayon EG, Khade RL, Powell DR, Zhang Y, Richter-Addo GB (2018) Lewis acid activation of the ferrous heme–NO fragment toward the N–N coupling reaction with NO to generate N2O. J Am Chem Soc 140:4204–4207
Zhu X, Burger M, Doane TA, Horwath WR (2013) Ammonia oxidation pathways and nitrifier denitrification are significant sources of N2O and NO under low oxygen availability. PNAS 110:6328–6333
Caranto JD, Vilbert AC, Lancaster KM (2016) Nitrosomonas europaea cytochrome P460 is a direct link between nitrification and nitrous oxide emission. PNAS 113:14704–14709
Vilbert AC, Caranto JD, Lancaster KM (2017) Influences of the heme–lysine crosslink in cytochrome P460 over redox catalysis and nitric oxide sensitivity. Chem Sci:DOI. https://doi.org/10.1039/c1037sc03450d
Shoun H, Fushinobu S, Jiang L, Kim S-W, Wagaki T (2012) Fungal denitrification and nitric oxide reductase cytochrome P450nor. Phil Trans R Soc B 367:1186–1194
McQuarters AB, Wirgau NE, Lehnert N (2014) Model complexes of key intermediates in fungal cytochrome P450 nitric oxide reductase (P450nor). Curr Op Chem Biol 19:82–89
Galloway JN, Townsend AR, Erisman JW, Bekunda M, Cai Z, Freney JR, Martinelli LA, Seitzinger SP, Sutton MA (2008) Transformation of the Nitrogen Cycle: recent trends, questions and potential solutions. Science 320:889–892
Duce RA, LaRoche J, Altieri K, Arrigo KR, Baker AR, Capone DG, Cornell S, Dentener F, Galloway J, Ganeshram RS, Geider RJ, Jickells T, Kuypers MM, Langlois R, Liss PS, Liu SM, Middelburg JJ, Moore CM, Nickovic S, Oschlies A, Pedersen T, Prospero J, Schlitzer R, Seitzinger S, Sorensen LL, Uematsu M, Ulloa O, Voss M, Ward B, Zamora L (2008) Impacts of atmospheric anthropogenic nitrogen on the open ocean. Science 320:893–897
Canfield DE, Glazer AN, Falkowski PG (2010) The evolution and future of earth’s nitrogen cycle. Science 330:192–196
Galloway JN, Leach AM, Bleeker A, Erisman JW (2013) A chronology of human understanding of the nitrogen cycle. Phil Trans R Soc B 368:20130120
Lehnert N, Coruzzi G, Hegg E, Seefeldt L, Stein L (2015) Feeding the world in the 21st century: grand challenges in the nitrogen cycle. National Science Foundation Workshop. https://www.nsf.gov/mps/che/workshops/nsf_nitrogen_report_int.pdf. Accessed June 2017
Ravishankara AR, Daniel JS, Portmann RW (2009) Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science 326:123–125
Wuebbles DJ (2009) Nitrous oxide: no laughing matter. Science 326:56–57
Eroy-Reveles A, Leung Y, Beavers CM, Olmstead MM, Mascharak PK (2008) Near-infrared light activated release of nitric oxide from designed photoactive manganese nitrosyls: strategy, design, and potential as NO donors. J Am Chem Soc 130:4447–4458
Koebke KJ, Pauly DJ, Lerner L, Liu X, Pacheco AA (2013) Does the oxidation of nitric oxide by oxyMyoglobin share an intermediate with the metMyoglobin-catalyzed isomerization of peroxynitrite? Inorg Chem 52:7623–7632
Koebke KJ, Waletzko MT, Pacheco AA (2016) Direct monitoring of the reaction between photochemically generated nitric oxide and Mycobacterium tuberculosis truncated hemoglobin N wild type and variant forms: an assessment of theoretical mechanistic predictions. Biochemistry 55:686–696
Drago RS, Paulik FE (1960) The reaction of nitrogen (II) oxide with diethylamine. J Am Chem Soc 82:96–98
Maragos CM, Morley D, Wink DA, Dunams TM, Saavedra JE, Hoffman A, Bove AA, Isaac L, Hrabie JA, Keefer LK (1991) Complexes of NO with nucleophiles as agents for the controlled biological release of nitric-oxide—vasorelaxant Effects. J Med Chem 34(11):3242–3247
Wanat A, Schneppensieper T, Karocki A, Stochel G, van Eldik R (2002) Thermodynamics and kinetics of RuIII(edta) as an efficient scavenger for nitric oxide in aqueous solution. J Chem Soc Dalton Trans 6:941–950
Chatterjee D, Mitra A, Sengupta A, Saha P, Chatterjee M (2006) [RuIII(Medtra)(H2O)] (medtra = N-methylethylenediaminetriacetate) complex—a highly efficient NO inhibitor with low toxicity. Inorg Chim Acta 359:2285–2290
Watanabe T, Honda K (1982) Measurement of the extinction coefficient of the methyl viologen cation radical and the efficiency of its formation by semiconductor photocatalysis. J Phys Chem 86:2617–2619
McGarry JM (2017) Interaction with nitric oxide of the Nitrosomonas europaea tetraheme protein cytochrome c 554, and two of its variants, in increasingly reducing environments. Ph.D., University of Wisconsin-Milwaukee, Dept. of Chemistry and Biochemistry, Milwaukee, WI
Koebke KJ (2015) Mechanistic study of heme protein-mediated nitric oxide dioxygenation using photolytically produced nitric oxide. Ph.D., University of Wisconsin-Milwaukee, Dept. of Chemistry and Biochemistry, Milwaukee, WI
Purwar N, McGarry JM, Kostera J, Pacheco AA, Schmidt M (2011) Interaction of nitric oxide with catalase: structural and kinetic analysis. Biochemistry 50:4491–4503
Cabail MZ, Moua V, Bae E, Meyer A, Pacheco AA (2007) Quantifying the photoinduced release of nitric oxide from N,N’-bis(carboxymethyl)-N,N’-dinitroso-1,4-phenylenediamine. Effect of reducing agents on the mechanism of the photoinduced reactions. J Phys Chem A 111(7):1207–1213. https://doi.org/10.1021/Jp065487y
Kostera J, Youngblut MD, Slosarczyk JM, Pacheco AA (2008) Kinetic and product distribution analysis of NO reductase activity in Nitrosomonas europaea hydroxylamine oxidoreductase. J Biol Inorg Chem 13(7):1073–1083. https://doi.org/10.1007/s00775-008-0393-4
Kostera J, McGarry JM, Pacheco AA (2010) Enzymatic interconversion of ammonia and nitrite: the right tool for the job. Biochemistry 49:8546–8553
Youngblut M, Judd ET, Srajer V, Sayyed B, Goelzer T, Elliott SJ, Schmidt M, Pacheco AA (2012) Laue crystal structure of Shewanella oneidensis cytochrome c nitrite reductase from a high-yield expression system. J Biol Inorg Chem 17:647–662
Youngblut M, Pauly DJ, Stein N, Walters D, Conrad JA, Moran GR, Bennett B, Pacheco AA (2014) Shewanella oneidensis cytochrome c nitrite reductase (ccNiR) does not disproportionate hydroxylamine to ammonia and nitrite, despite a strongly favorable driving force. Biochemistry 53:2136–2144
Arciero DM, Collins MJ, Haladjian J, Bianco P, Hooper AB (1991) Resolution of the 4 hemes of cytochrome-C554 from Nitrosomonas europaea by redox potentiometry and optical spectroscopy. Biochemistry 30(48):11459–11465
Pulcu GS, Elmore BL, Arciero DM, Hooper AB, Elliott SJ (2007) Direct electrochemistry of tetraheme cytochrome c 554 from Nitrosomonas europaea: redox cooperativity and gating. J Am Chem Soc 129:1838–1839
Press WH, Teukolsky SA, Vetterling WT, Flannery BP (2007) Numerical recipes the art of scientific computing, 3rd edn. Cambridge University Press, New York, pp 65–75
Henry ER, Hofrichter J (1992) Singular value decomposition: application to analysis of experimental data. In: Brand L, Johnson ML (eds) Meth enzymol, vol 210. Academic Press, San Diego, pp 129–192
Laverman LE, Wanat A, Oszajca J, Stochel G, Ford PC, van Eldik R (2001) Mechanistic studies on the reversible binding of nitric oxide to metmyoglobin. J Am Chem Soc 123(2):285–293. https://doi.org/10.1021/Ja001696z
Marritt SJ, Kemp GL, Xiaoe L, Durrant JR, Cheesman MR, Butt JN (2008) Spectroelectrochemical characterization of a pentaheme cytochrome in solution and as electrocatalytically active films on nanocrystalline metal-oxide electrodes. J Am Chem Soc 130:8588–8589
Enemark JH, Feltham RD (1974) Principles of structure, bonding and reactivity for metal nitrosyl complexes. Coord Chem Rev 13:339–406
Goldstein S, Czapski G (1995) Kinetics of nitric oxide autoxidation in aqueous solution in the absence and presence of various reductants. The nature of the oxidizing intermediates. J Am Chem Soc 117:12078–12084
Goldstein S, Czapski G (1996) Mechanism of the nitrosation of thiols and amines by oxygenated NO solutions: the nature of the nitrosating intermediates. J Am Chem Soc 118:3419–3425
Moore EG, Gibson QH (1976) Cooperativity in the dissociation of nitric oxide from hemoglobin. J Biol Chem 251:2788–2794
Hoshino M, Ozawa K, Seki H, Ford PC (1993) Photochemistry of nitric-oxide adducts of water-soluble iron(III) porphyrin and ferrihemoproteins studied by nanosecond laser photolysis. J Am Chem Soc 115(21):9568–9575
Hunt AP, Lehnert N (2015) Heme-nitrosyls: electronic structure implications for function in biology. Acc Chem Res 48:2117–2125
Praneeth VKK, Paulat F, Berto TC, George SD, Nather C, Sulok CD, Lehnert N (2008) Electronic structure of six-coordinate iron(III)-porphyrin NO adducts: the elusive iron(III)-NO(radical) state and its influence on the properties of these complexes. J Am Chem Soc 130:15288–15303
Ford PC, Lorkovic IM (2002) Mechanistic aspects of the reactions of nitric oxide with transition-metal complexes. Chem Rev 102(4):993–1017. https://doi.org/10.1021/Cr0000271
Wolak M, van Eldik R (2002) To be or not to be NO in coordination chemistry? A mechanistic approach. Coord Chem Rev 230(1–2):263–282 pii S0010-8545(01)00472-6
Lin R, Farmer PJ (2000) The HNO adduct of myoglobin: synthesis and characterization. J Am Chem Soc 122:2393–2394
Murugaeson RK, Fukuto JM, Miranda KM, Farmer PJ (2010) Reactions of HNO with heme proteins: new routes to HNO-heme complexes and insight into physiological effecs. Inorg Chem 49:6283–6292
Goodrich LE, Roy S, Alp EE, Zhao J, Hu MY, Lehnert N (2013) Electronic structure and biologically relevant reactivity of low-spin {Fe(NO)}8 porphyrin model complexes: new insight from a bis-picket fence porphyrin. Inorg Chem 52:7766–7780
Kurnikov IV, Ratner MA, Pacheco AA (2005) Redox equilibria in hydroxylamine oxidoreductase. Electrostatic control of electron redistribution in multielectron oxidative processes. Biochemistry 44(6):1856–1863. https://doi.org/10.1021/Bi048060v
Choi I-K, Liu Y, Feng DW, Paeng K-J, Ryan MD (1991) Electrochemical and spectroscopic studies of iron porphyrin nitrosyls and their reduction products. Inorg Chem 30:1832–1839
Lancon D, Kadish KM (1983) Electrochemical and spectral characterization of iron mono- and dinitrosyl porphyrins. J Am Chem Soc 105:5610–5617
Berto TC, Praneeth VKK, Goodrich LE, Lehnert N (2009) Iron-porphyrin NO complexes with covalently attached N-donor ligands: formation of a stable six-coordinate species in solution. J Am Chem Soc 131:17116–17126
Acknowledgements
Major funding for this work was provided by National Science Foundation Grants MCB-1121770, MCB-1330809, and MCB-1616824. The authors are indebted to Graham Moran and Nicholas Silvaggi of the University of Wisconsin-Milwaukee for use of their stopped-flow and FPLC chromatography systems, respectively, and to John Coates from the University of California-Berkeley for providing the WM3064 cells. JMM also wished to thank her colleagues in the laboratory, particularly Karl Koebke and Matthew Youngblut, for helpful discussions and much practical advice.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
McGarry, J.M., Pacheco, A.A. Upon further analysis, neither cytochrome c554 from Nitrosomonas europaea nor its F156A variant display NO reductase activity, though both proteins bind nitric oxide reversibly. J Biol Inorg Chem 23, 861–878 (2018). https://doi.org/10.1007/s00775-018-1582-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-018-1582-4