Abstract
Corynebacterium glutamicum has a long and successful history in the biotechnological production of l-lysine. Besides the adjustment of metabolic pathways, intracellular and extracellular transport systems are critical for the cellular metabolism of l-lysine or its by-products. Here, three amino acid transmembrane transporters, namely, GluE, BrnE/BrnF, and LysP, which are widely present in C. glutamicum strains, were each investigated by gene knockout. In comparison with that in the wild-type strain, the yield of l-lysine increased by 9.0%, 12.3%, and 10.0% after the deletion of the gluE, brnE/brnF, and lysP genes, respectively, in C. glutamicum 23,604. Moreover, the amount of by-product amino acids decreased significantly when the gluE and brnE/brnF genes were deleted. It was also demonstrated that there was no effect on the growth of the strain when the gluE or lysP gene was deleted, whereas the biomass of C. glutamicum WL1702 (ΔbrnE/ΔbrnF) in the fermentation medium was significantly reduced in comparison with that of the wild type. These results also provide useful information for enhancing the production of l-lysine or other amino acids by C. glutamicum.



taken from the fermentation medium every 4 h to measure the l-glutamate, l-lysine, glucose and OD600 production of C. glutamicum 23604 and C. glutamicumWL1701, symbols: filled square strain C. glutamicum 23604, hollow triangle strain C. glutamicum WL1701. The standard errors are shown as bars

taken from the fermentation medium every 4 h to measure the l-glutamate concentration of C. glutamicum 23604 and C. glutamicum WL1702, symbols: filled square strain C. glutamicum 23604, filled triangle strain C. glutamicum WL1702. The standard errors are shown as bars

taken from the fermentation medium every 4 h to measure the l-lysine, glucose and OD600 of C. glutamicum 23604 and C. glutamicum WL1702, symbols: filled square strain C. glutamicum 23604, filled triangle strain C. glutamicum WL1702. The standard errors are shown as bars

taken from the fermentation medium every 4 h to measure the l-lysine concentration of C. glutamicum 23604 and C. glutamicum WL1702, symbols: filled square strain C. glutamicum 23604, filled triangle strain C. glutamicum WL1702. The standard errors are shown as bars

taken from the fermentation medium every 4 h to measure the l-lysine, glucose and OD600 production of C. glutamicum 23604 and C. glutamicum WL1703, symbols: hollow square strain C. glutamicum 23604, filled triangle strain C. glutamicum WL1703. The standard errors are shown as bars

Similar content being viewed by others
Abbreviations
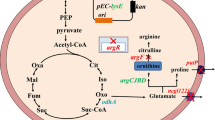
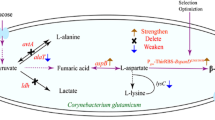
- AcCoA:
-
Acetyl coenzyme A
- Ala:
-
l-Alanine
- Asn:
-
l-Asparagine
- Asp:
-
l-Aspartate
- AspSa:
-
Aspartate semialdehyde
- Chor:
-
Chorismate
- Cit:
-
Citrate
- Fum:
-
Fumaric acid
- G6P:
-
Glucose 6-phosphate
- GDL:
-
Gluconolactone
- Glc:
-
Glucose
- Gln:
-
l-Glutamine
- Glu:
-
l-Glutamate
- Gluc:
-
d-Gluconic acid
- Gluc6P:
-
6-Phospho-d-gluconate
- Glx:
-
Glyoxylate
- Gly:
-
l-Glycine
- Hcys:
-
Homocysteine
- His:
-
l-Histidine
- Hser:
-
Homoserine
- ICit:
-
Isocitrate
- Ile:
-
l-Isoleucine
- αKG:
-
α-Ketoglutarate
- Kval:
-
2-Ketovaline
- Leu:
-
l-Leucine
- Lys:
-
l-Lysine
- Mal:
-
Malic acid
- OA:
-
Oxaloacetic acid
- Met:
-
l-Methionine
- Orn:
-
Ornithine
- PEP:
-
Phosphoenolpyruvate
- PG3:
-
Glycerate 3-phosphate
- Phe:
-
l-Phenylalanine
- Pro:
-
l-Proline
- PRPP:
-
5-Phospho-d-ribosylpyrophosphate
- Pyr:
-
Pyruvate
- R5P:
-
Ribulose 5-phosphate
- Ser:
-
l-Serine
- Suc:
-
Succinate
- SucCoA:
-
Succinate coenzyme A
- Thr:
-
l-Threonine
- Trp:
-
l-Tryptophan
- Tyr:
-
l-Tyrosine
- Val:
-
l-Valine
References
Bröer S, Krämer R (1990) Lysine uptake and exchange in Corynebacterium glutamicum. J Bacteriol 172:7241–7248. https://doi.org/10.1128/jb.172.12.7241-7248.1990
Chen C, Li Y, Hu J, Dong X, Wang X (2015) Metabolic engineering of Corynebacterium glutamicum ATCC13869 for l-valine production. Metab Eng 29:66–75. https://doi.org/10.1016/j.ymben.2015.03.004
Christian TT, Dietrich D, Brigitte B, Andreas B, Reinhard KM (2005) Characterization of methionine export in Corynebacterium glutamicum. J Bacteriol 187:3786. https://doi.org/10.1128/JB.187.11.3786-3794.2005
Hou X, Ge X, Wu D, Qian H, Zhang W (2012a) Improvement of l-valine production at high temperature in Brevibacterium flavum by overexpressing ilvEBNrC genes. J Ind Microbiol Biotechnol 39:63–72. https://doi.org/10.1007/s10295-011-1000-1
Hou X, Zhang Y, Qian H, Zhang W (2012b) l-Valine production with minimization of by-products’ synthesis in Corynebacterium glutamicum and Brevibacterium flavum. Amino Acids 43:2301–2311. https://doi.org/10.1007/s00726-012-1308-9
Ikeda M (2017) Lysine fermentation: history and genome breeding. Adv Biochem Eng Biotechnol 159:73–102. https://doi.org/10.1007/10_2016_27
Jun N, Seiko H, Hisao I, Masaaki W (2007) Mutations of the Corynebacterium glutamicum NCgl1221 gene, encoding a mechanosensitive channel homolog, induce l-glutamic acid production. Appl Environ Microbiol 73:4491–4498. https://doi.org/10.1128/AEM.02446-06
Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A et al (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J Biotechnol 104(1–3):5–25. https://doi.org/10.1016/s0168-1656(03)00154-8
Katheline H (2013) Investigating the appropriate mode of expressing lysine requirement of fish through non-linear mixed model analysis and multilevel analysis. Br J Nutr 109:1013–1021. https://doi.org/10.1017/S0007114512002863
Kennerknecht N, Sahm H, Yen MR, Pátek M, Eggeling L (2002) Export of l-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family. J Bacteriol 184:3947–3956. https://doi.org/10.1128/JB.184.14.3947-3956.2002
Koffas M, Stephanopoulos G (2005) Strain improvement by metabolic engineering: lysine production as a case study for systems biology. Curr Opin Biotechnol 16:361–366. https://doi.org/10.1016/j.copbio.2005.04.010
Lange C, Mustafi N, Frunzke J, Kennerknecht N, Wessel M, Bott M, Wendisch VF (2012) Lrp of Corynebacterium glutamicum controls expression of the brnEF operon encoding the export system for l-methionine and branched-chain amino acids. J Biotechnol 158:231–241. https://doi.org/10.1016/j.jbiotec.2011.06.003
Marin K, Krämer R (2007) Amino acid transport systems in biotechnologically relevant bacteria. Amino Acid 5:289–325. https://doi.org/10.1007/7171_2006_069
Nakamura J, Hirano S, Ito H, Wachi M (2007) Mutations of the Corynebacterium glutamicum NCgl1221 gene, encoding a mechanosensitive channel homolog, induce l-glutamic acid production. Appl Environ Microbiol 73:4491–4498. https://doi.org/10.1128/AEM.02446-06
Nicole K, Hermann S, Ming-Ren Y, Miroslav P Jr, Saier MH, Lothar E (2002) Export of l-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family. J Bacteriol 184:3947–3956. https://doi.org/10.1128/JB.184.14.3947-3956.2002
Nji E, Li D, Doyle DA, Caffrey M (2014) Cloning, expression, purification, crystallization and preliminary X-ray diffraction of a lysine-specific permease from Pseudomonas aeruginosa. Acta Crystallogr A 70:1362–1367. https://doi.org/10.1107/S2053230X14017865
Qin T, Hu X, Hu J, Wang X (2014) Metabolic engineering of Corynebacterium glutamicum strain ATCC13032 to produce l-methionine. Biotechnol Appl Biochem 62(4):563–573. https://doi.org/10.1117/12.2061040
Ruan Y, Zhu L, Qi L (2015) Improving the electro-transformation efficiency of Corynebacterium glutamicum by weakening its cell wall and increasing the cytoplasmic membrane fluidity. Biotechnol Lett 37:2445–2452. https://doi.org/10.1007/s10529-015-1934-x
Seep-Feldhaus AH, Kalinowski J, Pühler A (2010) Molecular analysis of the Corynebacterium glutamicum lysl gene involved in lysine uptake. Mol Microbiol 5:2995–3005. https://doi.org/10.1111/j.1365-2958.1991.tb01859.x
Shukuo K, Shigezo U, Masakazu S (2004) Studies on the amino acid fermentation. Part 1. Production of l-glutamic acid by various microorganisms. J Gen Appl Microbiol 50:331–343. https://doi.org/10.1002/jgm.614
Vrljic M, Sahm H, Eggeling L (1997a) A new type of transporter with a new type of cellular function: l-lysine export from Corynebacterium glutamicum. Mol Microbiol 22:815–826. https://doi.org/10.1046/j.1365-2958.1996.01527.x
Wittmann C, Becker J (2007) The l-lysine story: from metabolic pathways to industrial production. Amino Acid. https://doi.org/10.1007/7171_2006_089
Yamashita C, Hashimoto KI, Kumagai K, Maeda T, Wachi M (2013) l-Glutamate secretion by the N-terminal domain of the Corynebacterium glutamicum NCgl1221 mechanosensitive channel. Biosci Biotechnol Biochem 77:1008–1013. https://doi.org/10.1271/bbb.120988
Yao W, Deng X, Liu M, Zheng P, Sun Z, Zhang Y (2009) Expression and localization of the Corynebacterium glutamicum NCgl1221 protein encoding an l-glutamic acid exporter. Microbiol Res 164:680–687. https://doi.org/10.1016/j.micres.2009.01.001
Yoshitaka N, Kenjiro Y, Hidetoshi I (2012) A gain-of-function mutation in gating of Corynebacterium glutamicum NCgl1221 causes constitutive glutamate secretion. Appl Environ Microbiol 78:5432–5434. https://doi.org/10.1128/AEM.01310-12
Acknowledgements
This work was supported by the National Science Foundation of China (31801527), Focus on Research and Development Plan in Shandong Province (2019JZZY011003, 2018YFJH0401, 2016CYJS07A01), Taishan industry leading talent (tscy20180103), Shandong Provincial Natural Science Foundation (ZR2016CB04), Major Program of National Natural Science Foundation of Shandong (ZR2017ZB0208) and Cultivation Project of Shandong Synthetic Biotechnology Innovation Center (sdsynbio-2018-PY-02).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
Communicated by S. Stuchlík.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xiao, J., Wang, D., Wang, L. et al. Increasing l-lysine production in Corynebacterium glutamicum by engineering amino acid transporters. Amino Acids 52, 1363–1374 (2020). https://doi.org/10.1007/s00726-020-02893-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-020-02893-6