Abstract
H9N2 avian influenza viruses (AIVs) affect both poultry and humans on a global level, and they are especially prevalent in Egypt. In this study, we sequenced the entire genome of AIV H9N2 isolated from chickens in Egypt in 2021, using next-generation sequencing (NGS) technology. Phylogenetic analysis of the resulting sequences showed that the studied strain was generally monophyletic and grouped within the G1 sublineage of the Eurasian lineage. Four segments (polymerase basic 2 [PB2], polymerase basic 1 [PB1], polymerase acidic [PA], and non-structural [NS]) were related to Egyptian genotype II, while the nucleoprotein (NP), neuraminidase (NA), matrix (M), and haemagglutinin (HA) segments were related to Egyptian genotype I. Molecular analysis revealed that HA protein contained amino acid residues (191H and 234L) that suggested a predilection for attaching to human-like receptors. The antigenic sites of HA had two nonsynonymous mutations: V194I at antigenic site A and M40K at antigenic site B. Furthermore, the R403W and S372A mutations, which have been observed in H3N2 and H2N2 strains that caused human pandemics, were found in the NA protein of the detected strain. The internal proteins contained virulence markers: 504V in the PB2 protein, 622G, 436Y, 207K, and 677T in the PB1 protein, 127V, 550L, and 672L in PA protein, and 64F and 69P in the M protein. These results show that the detected strain had undergone intrasubtype reassortment. Furthermore, it contains changes in the viral proteins that make it more likely to be virulent, raising a question about the tendency of AIV H9N2 to become highly pathogenic in the future for both poultry and humans.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In 1966, turkeys in the United States were the source of an avian influenza virus (AIV) H9N2 strain [1], which then split into Eurasian and American lineages [2]. According to Lee and Song [3], the Eurasian group has been further divided into the Korean lineage, the Y280 lineage, and the G1-like lineage, with the G1-like lineage being the most prevalent of these [3, 4].
In 2010, H9N2 G1-like lineages in Egypt were reported [5,6,7], where the virus subsequently became prevalent in domestic poultry. Based on phylogenetic analysis, Egyptian H9N2 viruses are classified into two genotypes: genotype I, which was found in Egyptian poultry between 2010 and 2013, and genotype II, which emerged in 2014 as a result of reassortment between AIV H9N2 G1 and AIV H9N2 Eurasian strains from wild birds [8, 9].
Influenza A virus is a member of the family Orthomyxoviridae. Its genome consists of eight dsRNA segments that encode at least 10 viral proteins and are arranged in ascending order as follows: polymerase basic protein 2 (PB2), polymerase basic protein 1 (PB1), polymerase acidic protein (PA), hemagglutinin (HA), nucleoprotein (NP), neuraminidase (NA), matrix proteins (M1 and M2), and non-structural proteins (NS1 and NS2 or nuclear export protein [NEP]) [10].
Despite being categorized as low-pathogenicity avian influenza viruses (LPAIs), H9 viruses have been known to exhibit a highly pathogenic phenotype in both laboratory and field settings due to mutations that increase virulence and lethality [11, 12]. Due to their zoonotic potential, H9N2 viruses also represent a threat to human health worldwide, especially since multiple human infections have already been reported [13,14,15,16]. Currently, the capability of H9N2 viruses to contribute their genes to other AIVs that can cross the species barrier to infect humans, such as zoonotic H5N6 [17] and H7N9 [18], is of particular concern worldwide.
Numerous investigations have been carried out to identify viral factors that are linked to increased pathogenicity, virulence, and transmissibility. Specifically, genetic changes associated with airborne transmissibility and adaptation to replication in mammalian hosts are of particular importance [19]. Changes that affect viral entry, viral polymerase activity, and the host response are the main factors determining virulence, and they also affect the efficiency of infection and spread to new hosts [20].
Influenza viruses use both reassortment and individual mutations to adapt to their hosts [21]. Since their first discovery in 1966, H9N2 viruses have undergone evolution and reassortment with other viral subtypes, resulting in a significant increase in their genetic diversity [22]. According to Pusch and Suarez [23], only the G1 and Y280/G9 lineages of H9N2 viruses have been verified to be infectious in humans.
The current investigation was carried out in order to trace the evolution of Egyptian H9N2 viruses and to determine whether new reassortment events that increase the zoonotic potential and virulence of these viruses are likely to occur.
Materials and methods
Sample collection
One hundred oropharyngeal and cloacal swabs were collected from birds with respiratory manifestations from Egyptian commercial poultry farms (chickens, ducks, and turkeys) with a 15–25% mortality rate during the period from 2020 to 2021. A total of 5 to 10 individual oropharyngeal and/or cloacal swabs collected from each farm were pooled together and treated as one sample. The samples were obtained from 10 Egyptian governorates (Bahira, Dakahlia, Damietta, Giza, Gharbia, Ismailia, Kafr El Sheikh, Menia, Menoufia, and Sharkia). The epidemiological data of the collected samples are provided in Supplementary Table S1.
Molecular detection and virus isolation
Viral RNA was extracted using a QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions in a class 2 biological safety cabinet (SterilGARD, USA). The RNA purity was measured spectrophotometrically using a NanoDrop 2000/2000c instrument (Thermo Fisher Scientific, Waltham, MA, USA). The purity of the extracted viral RNA (A260/A280 ratio) ranged from 1.7 to 2. The extracted RNAs were subjected to quantitative reverse transcription polymerase chain reaction (RT-qPCR) to test for the presence of the influenza A virus matrix (M) gene [24].
Following the standard procedures of the World Organization for Animal Health (OIE) diagnostic handbook, positive samples were inoculated into the allantoic cavities of 9- to 11-day-old specific-pathogen-free embryonated chicken eggs. About 48 hours after inoculation, allantoic fluids were collected, viral RNA was extracted, and RT-PCR was performed for detection of the H5, H9, N8, and N2 genes [25, 26].
Whole-genome sequencing
Amplicons of each genome segment of an isolate of the H9N2 virus were generated using a SuperScript IV One-Step RT-PCR Kit with Platinum SuperFi DNA Polymerase. The amplicons were sequenced using Ion Torrent next-generation sequencing (NGS) technology (the Ion PGM System with an Ion 316 chip). The reads were analyzed using the Geneious Prime work package (Biomatters, Auckland, New Zealand) as follows: The primer sequences were removed from the raw reads using the “Trim Ends” Geneious Prime plugin. Next, the trimmed reads were mapped using bowtie2, implemented in Geneious Prime, against a reference whole-genome sequence. The resulting sequences were submitted to the GenBank database. The strain was designated A/chicken/Egypt/Menoufia/2021, and the accession numbers are listed in Supplementary Table S2. Whole-genome sequences of H9N2 viruses were downloaded from the NCBI database, and a multiple alignment was made for each genome segment using the Clustal W multiple alignment accessory application in BioEdit software version 7.2.5 (BioEdit Company, Manchester, UK).
Phylogenetic profiling and molecular characterization
The sequence alignments were used to create a phylogenetic tree by the maximum-likelihood method in MEGA11 software [27], employing the general time-reversible (GTR) nucleotide substitution model. The robustness of the tree branches was estimated using 1000 bootstrap replicates. The antigenic sites and the genetic signature markers associated with virulence, host tropism, enhanced replication, and drug resistance were identified using the aligned amino acid sequences. BioEdit version 7.2.5 was used to compare nucleotide and protein sequences. Potential glycosylation sites were identified using the NetN-Glyc 1.0 server [28]. SWISS-MODEL was used to model the HA protein structures of the Egyptian H9N2 virus and the parental Egyptian AIV H9N2 virus (A/chicken/Egypt/S4456B/2011) [29], and their structures were visualized using PyMOL 1.1 (DeLano Scientific LLC).
Results
Sample screening and virus detection
RT-qPCR testing for the avian influenza virus M gene revealed that the virus was present on 23 farms (23%). Three of the positive samples, from farms in Menoufia governorate, contained subtype H9N2. The chickens on these three farms had been vaccinated with a killed H9 vaccine when they were three days old and with an H5 (clade 2.2.1) vaccine when they were eight days old. The mortality rate ranged from 20–22%. Amplification curves and conventional PCR results are shown in Supplementary Figures S1a and b, S2, and S3.
Phylogenetic profiling and sequence similarity of H9N2 isolates
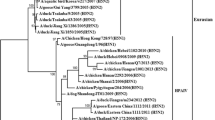
Phylogenetic analysis based on the surface genes (HA and NA) of the AIV H9N2 virus showed that it belonged to the G1 sublineage of the Eurasian lineage. Our isolate was monophyletic with recent Egyptian AIV H9N2 isolates in the GenBank database, such as A/chicken/Egypt/A19610/2021 and A/chicken/Egypt/N19766D/2021 (Fig. 1), with 99% nucleotide sequence identity in the HA and NA genes (Table 1). However, it was was relatively distant from the parental Egyptian AIV H9N2 virus (A/chicken/Egypt/S4456B/2011) (Fig. 1), with 93% nucleotide sequence identity in the HA gene and 93.5% identity in the NA gene (Table 1). The internal genes (PB2, PB1, PA, NP, M, and NS) of our AIV H9N2 isolate grouped with the G1 sublineage of the Eurasian lineage (Fig. 1), with 99% nucleotide sequence identity to recently published PB1, PB2, M, NP, and NS sequences of Egyptian AIV H9N2 viruses and 97% identity in the PA gene (Table 1). The parental Egyptian AIV H9N2 virus (A/chicken/Egypt/S4456B/2011) was 86%, 89%, 89.55, 94%, 96%, and 89.5% identical in the PB2, PB1, Pa, NP, M, and NS gene, respectively (Table 1). The PB2, PB1, PA, and NS genes showed a close relationship to isolates of Egyptian genotype II (Fig. 1), while the HA, NP, NA, and M genes were more closely related to isolates of Egyptian genotype I (Fig. 1).
Molecular characterization
The Egyptian AIV H9N2 isolate from this study has a monobasic motif (PARSSRGLFG) at the cleavage site of the HA protein, which resembles those found in low-pathogenicity AIVs. It also has several amino acid residues in the receptor-binding site (RBS) of the HA that are associated with a preference for binding to human-like α2,6 sialic acid (191H, 232N, 234L, 235I, and 236G) (Fig. 2). In addition, when compared with the parental Egyptian H9N2 virus, the studied strain was found to have gained two substitutions in antigenic sites: V194I in antigenic site A and M40K in antigenic site B (Fig. 2). Moreover, the HA had seven potential N-linked glycosylation sites at positions 29 (NSTE), 82 (NPSC), 105 (NGTC), 141 (NVTY), 298(NSTL), 305(NISK), and 492 (NGTY).
Genetic analysis of the region encoding the three loops that form the hemadsorption site on the NA gene revealed the presence of three amino acid substitutions: S372A (N2 numbering) in the first loop and I402N and R403W (N2 numbering) in the second loop (Fig. 3) when compared with the A/Quail/Hong Kong/G1/97 prototype (Table 2). In addition, the NA gene has six strong N-linked glycosylation sites at positions 44 (NTST), 61 (NITE), 69 (NGTI), 86 (NWSK), 146 (NGTI), and 234 (NGTC). The polymerase complex (PB2, PB1, and PA) showed the presence of ten markers related to enhanced virulence: V at amino acid position 504 in PB2, 13P, 436Y, 207K, 677T, and 622G in PB1, V at position 127, D at position 383 and L at positions 550 and 672 in PA (Table 3), and three mammalian preference markers: K318R and M64T substitutions in PB2 and D at position 382 in PA.
The M protein was found to have two virulence markers – F at amino acid position 64 and P at position 69 (Table 3) – and three markers associated with mammalian host-specificity: N at position 20, V at position 28, and F at position 55. The NS protein was found to have S at amino acid position 42 and A at position 149, both of which are virulence markers (Table 3). The NP protein was found to have Q at amino acid position 398, which is a unique marker for mammalian host specificity.
Discussion
Since its identification in Egypt in 2010, the AI H9N2 virus has become prevalent due to its simultaneous circulation alongside clade 2.2.1 H5N1 viruses, which were endemic in the region [30]. In 2017, H5N8 was isolated in Egypt for the first time, and the cocirculation of H9N2 and H5N8 increased the likelihood that reassortment between these two subtypes would result in the appearance of new viruses with pandemic capability. For instance, a recent reassortant (H5N2) between the Egyptian H5N8 and H9N2 viruses has been reported [31, 32]. Furthermore, the zoonotic potential of H9N2 viruses has already been established, with at least 72 verified cases in humans [16].
In the current study, the whole genome sequence of an Egyptian AIV H9N2 virus was determined to evaluate its phylogenetic relationships and to identify molecular genetic markers related to virulence, pathogenicity, and mammalian host preference that were being carried by circulating AIV H9N2 viruses during 2021.
Phylogenetic analysis based on the HA gene showed that the H9N2 isolate A/chicken/Egypt/Menoufia/2021 is closely related to members of a G1-like lineage of H9N2 viruses that were isolated previously in Egypt [9]. Furthermore, a genotype replacement was observed, with phylogenetic analysis revealing that four segments (PB2, PB1, PA, and NS) were associated with genotype II. In contrast, the segments HA, NA, M, and NP were found to be related to those of genotype I viruses. The HA protein in the identified H9N2 strain was predicted to have the monobasic cleavage motif “PARSSRGLF” in the HA1-HA2 connecting peptide. Proteolytic cleavage activation at this site plays a critical role in the viral life cycle [33,34,35]. Compared with the parental Egyptian AIV H9N2 virus, the isolate from this study has gained two substitutions in antigenic sites: a V194I substitution in antigenic site A and an M40K substitution in antigenic site B. This suggests that antigenic changes have occurred in circulating AIV H9N2 viruses as a result of vaccine failure, illustrating the need for continual updating of commercially used vaccines to match the circulating strains. The H9N2 strain from this study has the 191H and 234L variations, which is associated with a change in the HA preference from avian α-2,3 sialic acid (SA) receptors to human α-2,6 SA receptors, which suggests the potential for binding to human respiratory epithelial cells, as reported by Sorrell et al. [36]. Matrosovich et al. reported that the 234L variation is characteristic of human pandemic AIV H2 and H3 subtypes. Additionally, the 191H variation is associated with enhanced replication in human respiratory cell cultures and a preference for binding to receptors on human respiratory cells [37,38,39]. A higher binding affinity of the strain under investigation to human-like receptors is further suggested by the presence of numerous substitutions in the RBS. Genetic examination of the HA sequence revealed that the Egyptian H9N2 strain had seven potential N-linked glycosylation sites, at positions 29 (NSTE), 82 (NPSC), 105 (NGTC), 141 (NVTY), 298 (NSTL), 305 (NISK), and 492 (NGTY). These sites play a crucial role in protein folding, trafficking, pH stability, receptor binding potential, infectivity, and cell-associated host immunological reactions [8, 32, 40]. The N-linked glycosylation site at position 82 has not been seen in recently isolated Egyptian AIV H9N2 viruses. Variations in the glycosylation pattern can have an impact on pathogenicity and the affinity and specificity of receptor binding [9, 41, 42].
Previous studies have shown that the length of the stalk, the positions of N-glycosylation sites, and residues in the enzyme active site are major molecular determinants of the functional activities of NA and that hemadsorption (sialic acid binding) enhances the catalytic efficiency of NA, and therefore, modifications in these locations could potentially affect the specificity of the host receptor and sialic acid binding [42]. In the current study, no stalk deletion was observed in the H9N2 isolate, but three amino acid substitutions were observed in the hemadsorption sites in comparison to the A/Quail/Hong Kong/G1/97 prototype: S372A, I402N, and R403W (N2 numbering). The S372A and R403W substitutions have been shown to enhance the capability of the virus to overcome species barriers and adapt to mammalian hosts. Notably, these substitutions have been observed in the H2N2 and H3N2 subtypes, contributing to pandemics in the human population [42,43,44]. In addition, the H9N2 strain in this study has six potential N-linked glycosylation sites in the NA gene. This pattern of glycosylation facilitates the cleavage of NA by cellular proteases, which in turn facilitates the spread of infection [9].
The internal proteins (PB2, PB1, PA, NP, M, and NS) of avian influenza virus also influence host tropism and pathogenicity [45, 46], Mutations in the replication complex genes, (PB2, PB1, and PA) have the potential to increase the viral replication rate [47]. In the present study, nine markers related to enhanced polymerase activity and increased virulence were found in components of the polymerase complex: 504V in PB2 [48], 13P, 436Y, 207K, 677T, and 622G in PB1 [49], and 127V, 550L, and 672L in PA [5, 45]. The PA gene plays a major role in the ability of the virus to adapt to new hosts [50, 51]. The PA protein of the studied isolate retained a D residue at position 383, which is present in avian influenza viruses and the G1 prototype strain. This residue may facilitate the crossing of species barriers, as it is linked to increased polymerase activity in avian and mammalian cell lines [52]. The combination of these mutations may enhance polymerase activity. NP plays several roles in the AIV life cycle and its pathogenicity, replication ability, and infectivity in mammals [53]. In this study, the NP protein had the amino acid residue Q at position 398, which is a unique marker of mammalian host preference. A number of variations associated with host tropism and the immune response were found in the M1 and M2 proteins [54, 55]. The M2 protein of our isolate contains markers related to virulence and mammalian host preference: the 64S and 69P variations, which are associated with increased virulence [5], and the 20N, 28V, and 55F variations, which are associated with mammalian host preference and therefore mammalian transmission and human cases [56]. The NS1 protein of our isolate has a PDZ ESEI (227–230) C-terminal motif, which is specific for avian species and is considered a virulence marker [56, 57]. In addition, the NS1 protein has the amino acid residue S at position 42 and A at position 149, both of which have been linked to elevated virulence and more-efficient viral replication in mammalian cells [58, 59].
Our study was limited by the number of sequenced strains and was confined to 10 Egyptian governorates. Thus, it is necessary to conduct additional research on larger numbers of samples to track the evolution of these strains in the field and evaluate their zoonotic potential. Also, an assessment of commercially used vaccines is recommended, and vaccines should be updated periodically. Further research employing animal models is necessary to examine the pathogenicity of the current H9N2 strains in Egypt.
Conclusion
We determined the whole genome sequence of an AIV H9N2 virus identified in broiler chickens showing respiratory signs in an outbreak with 22% mortality. Phylogenetic analysis revealed that this virus is related to members of the G1 sublineage of the Eurasian group. The studied virus had undergone intra-subtype reassortment, with four segments inherited from Egyptian genotype 1, while the other four segments were inherited from Egyptian genotype 2. This strain showed a number of non-synonymous mutations that are molecular markers of increased virulence, antigenic variability, and affinity for human-like receptors. Interestingly, one substitution was related to H2 and H3 subtypes. The combination of the detected markers indicates the continuous evolution of AI H9N2 in the field and highlights its potential to become highly pathogenic in both poultry and humans.
References
Homme PJ, Easterday BC (1970) Avian influenza virus infections. I. Characteristics of influenza A-turkey-Wisconsin-1966 virus. Avian Dis 14(1):66–74 PMID: 4314007
Banks J, Speidel EC, Harris PA, Alexander DJ (2000) Phylogenetic analysis of influenza A viruses of H9 haemagglutinin subtype. Avian Pathol 29(4):353–359. https://doi.org/10.1080/03079450050118485
Lee DH, Song CS (2013) H9N2 avian influenza virus in Korea: evolution and vaccination. Clin Exp Vaccine Res 2(1):26–33. https://doi.org/10.7774/cevr.2013.2.1.26
Shahsavandi S, Salmanian AH, Ghorashi SA, Masoudi S, Ebrahimi MM (2012) Evolutionary characterization of hemagglutinin gene of H9N2 influenza viruses isolated from Asia. Res Vet Sci 93(1):234–239. https://doi.org/10.1016/j.rvsc.2011.07.033
Arafa A, Suarez D, Kholosy SG, Hassan MK, Nasef S, Selim A, Dauphin G, Kim M, Yilma J, Swayne D, Aly MM (2012) Evolution of highly pathogenic avian influenza H5N1 viruses in Egypt indicating progressive adaptation. Arch Virol 157(10):1931–1947. https://doi.org/10.1007/s00705-012-1385-9
El-Zoghby EF, Arafa AS, Hassan MK, Aly MM, Selim A, Kilany WH, Selim U, Nasef S, Aggor MG, Abdelwhab EM, Hafez HM (2012) Isolation of H9N2 avian influenza virus from bobwhite quail (Colinus virginianus) in Egypt. Arch Virol 157(6):1167–1172. https://doi.org/10.1007/s00705-012-1269-z
Kandeil A, El-Shesheny R, Maatouq AM, Moatasim Y, Shehata MM, Bagato O, Rubrum A, Shanmuganatham K, Webby RJ, Ali MA, Kayali G (2014) Genetic and antigenic evolution of H9N2 avian influenza viruses circulating in Egypt between 2011 and 2013. Arch Virol 159(11):2861–2876. https://doi.org/10.1007/s00705-014-2118-z
Kandeil A, El-Shesheny R, Maatouq A, Moatasim Y, Cai Z, McKenzie P, Webby R, Kayali G, Ali MA (2017) Novel reassortant H9N2 viruses in pigeons and evidence for antigenic diversity of H9N2 viruses isolated from quails in Egypt. J Gen Virol 98(4):548–562. https://doi.org/10.1099/jgv.0.000657
El Sayes M, Kandeil A, Moatasim Y, El Taweel A, Rubrum A, Kutkat O, Kamel MN, Badra R, Barakat AB, McKenzie PP, El-Shesheny R, Webby RJ, Kayali G, Ali MA (2022) Insights into Genetic Characteristics and Virological Features of Endemic Avian Influenza A (H9N2) Viruses in Egypt from 2017–2021. Viruses 14(7):1484. https://doi.org/10.3390/v14071484
Mostafa A, Abdelwhab EM, Mettenleiter TC, Pleschka S (2018) Zoonotic Potential of Influenza A Viruses: A Comprehensive Overview. Viruses 10(9):497. https://doi.org/10.3390/v10090497
Nili H, Asasi K (2003) Avian influenza (H9N2) outbreak in Iran. Avian Dis 47(3 Suppl):828–831. https://doi.org/10.1637/0005-2086-47.s3.828
Zhang Y, Guo X, Qi J, Liu L, Wang J, Xu S, Wang J, Yin Y (2014) Complete Genome Sequence of an H9N2 Influenza Virus Lethal to Chickens. Genome Announc 2(6):e00929–e00914. https://doi.org/10.1128/genomeA.00929-14
Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM, Lim W, Webster RG, Yuen KY, Peiris JS, Guan Y (2005) Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol 43(11):5760–5767. https://doi.org/10.1128/JCM.43.11.5760-5767.2005
Khan SU, Anderson BD, Heil GL, Liang S, Gray GC (2015) A systematic review and meta-analysis of the seroprevalence of influenza A (H9N2) infection among humans. J Infect Dis 212(4):562–569. https://doi.org/10.1093/infdis/jiv109
Peacock THP, James J, Sealy JE, Iqbal M (2019) A Global Perspective on H9N2 Avian Influenza Virus. Viruses 11(7):620. https://doi.org/10.3390/v11070620
WHO (World Health Organization) (2022) Avian Influenza Weekly Update Number 836; WHO: Geneva, Switzerland
Yang L, Zhu W, Li X, Bo H, Zhang Y, Zou S, Gao R, Dong J, Zhao X, Chen W, Dong L, Zou X, Xing Y, Wang D, Shu Y (2017) Genesis and Dissemination of Highly Pathogenic H5N6 Avian Influenza Viruses. J Virol 91(5):e02199–e02116. https://doi.org/10.1128/JVI.02199-16
Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung CY, Chen X, Li L, Hong W, Chai Y, Zhou L, Liang H, Ou Z, Liu Y, Farooqui A, Kelvin DJ, Poon LL, Smith DK, Pybus OG, Leung GM, Shu Y, Webster RG, Webby RJ, Peiris JS, Rambaut A, Zhu H, Guan Y (2013) The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502(7470):241–244. https://doi.org/10.1038/nature12515
Zhang YH, Zhao Y, Li N, Peng YC, Giannoulatou E, Jin RH, Yan HP, Wu H, Liu JH, Liu N, Wang DY, Shu YL, Ho LP, Kellam P, McMichael A, Dong T (2013) Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat Commun 4:1418. https://doi.org/10.1038/ncomms2433
Liu T, Wang Y, Tan TJC, Wu NC, Brooke CB (2022) The evolutionary potential of influenza A virus hemagglutinin is highly constrained by epistatic interactions with neuraminidase. Cell Host Microbe 30(10):1363–1369e4. https://doi.org/10.1016/j.chom.2022.09.003
Neverov AD, Lezhnina KV, Kondrashov AS, Bazykin GA (2014) Intrasubtype reassortments cause adaptive amino acid replacements in H3N2 influenza genes. PLoS Genet 10(1):e1004037. https://doi.org/10.1371/journal.pgen.1004037
Dong G, Luo J, Zhang H, Wang C, Duan M, Deliberto TJ, Nolte DL, Ji G, He H (2011) Phylogenetic diversity and genotypical complexity of H9N2 influenza A viruses revealed by genomic sequence analysis. PLoS ONE 6(2):e17212. https://doi.org/10.1371/journal.pone.0017212
Pusch EA, Suarez DL (2018) The Multifaceted Zoonotic Risk of H9N2 Avian Influenza. Vet Sci 5(4):82. https://doi.org/10.3390/vetsci5040082
Spackman E, Senne DA, Bulaga LL, Myers TJ, Perdue ML, Garber LP, Lohman K, Daum LT, Suarez DL (2003) Development of real-time RT-PCR for the detection of avian influenza virus. Avian Dis 47(3 Suppl):1079–1082. https://doi.org/10.1637/0005-2086-47.s3.1079
Lee MS, Chang PC, Shien JH, Cheng MC, Shieh HK (2001) Identification and subtyping of avian influenza viruses by reverse transcription-PCR. J Virol Methods 97(1–2):13–22. https://doi.org/10.1016/s0166-0934(01)00301-9
Tsukamoto K, Ashizawa T, Nakanishi K, Kaji N, Suzuki K, Shishido M, Okamatsu M, Mase M (2009) Use of reverse transcriptase PCR to subtype N1 to N9 neuraminidase genes of avian influenza viruses. J Clin Microbiol 47(7):2301–2303. https://doi.org/10.1128/JCM.02366-08
Tamura K, Stecher G, Kumar S (2021) MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 38(7):3022–3027. https://doi.org/10.1093/molbev/msab120
Gupta R, Jung E, Brunak S (2004) Prediction of N-glycosylation sites in human proteins
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22(2):195–201. https://doi.org/10.1093/bioinformatics/bti770
Kayali G, Kandeil A, El-Shesheny R, Kayed AS, Gomaa MM, Maatouq AM, Shehata MM, Moatasim Y, Bagato O, Cai Z, Rubrum A, Kutkat MA, McKenzie PP, Webster RG, Webby RJ, Ali MA (2014) Active surveillance for avian influenza virus, Egypt, 2010–2012. Emerg Infect Dis 20(4):542–551. https://doi.org/10.3201/eid2004.131295
Nagy A, Mettenleiter TC, Abdelwhab EM (2017) A brief summary of the epidemiology and genetic relatedness of avian influenza H9N2 virus in birds and mammals in the Middle East and North Africa. Epidemiol Infect 145(16):3320–3333. https://doi.org/10.1017/S0950268817002576
Hassan KE, King J, El-Kady M, Afifi M, Abozeid HH, Pohlmann A, Beer M, Harder T (2020) Novel Reassortant Highly Pathogenic Avian Influenza A(H5N2) Virus in Broiler Chickens, Egypt. Emerg Infect Dis 26(1):129–133. https://doi.org/10.3201/eid2601.190570
Callan RJ, Hartmann FA, West SE, Hinshaw VS (1997) Cleavage of influenza A virus H1 hemagglutinin by swine respiratory bacterial proteases. J Virol 71(10):7579–7585. https://doi.org/10.1128/JVI.71.10.7579-7585.1997
Zhirnov OP, Konakova TE, Wolff T, Klenk HD (2002) NS1 protein of influenza A virus down-regulates apoptosis. J Virol 76(4):1617–1625. https://doi.org/10.1128/jvi.76.4.1617-1625.2002
Bertram S, Glowacka I, Steffen I, Kühl A, Pöhlmann S (2010) Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev Med Virol 20(5):298–310. https://doi.org/10.1002/rmv.657
Sorrell EM, Wan H, Araya Y, Song H, Perez DR (2009) Minimal molecular constraints for respiratory droplet transmission of an avian-human H9N2 influenza A virus. Proc Natl Acad Sci U S A 106(18):7565–7570. https://doi.org/10.1073/pnas.0900877106
Matrosovich MN, Gambaryan AS, Klenk HD (2008) Receptor specificity of influenza viruses and its alteration during interspecies transmission. Avian influenza 27:134–155 Karger Publishers. https://doi.org/10.1159/000151617
Wan H, Perez DR (2007) Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J Virol 81(10):5181–5191. https://doi.org/10.1128/JVI.02827-06
Wan XF, Ozden M, Lin G (2008) Ubiquitous reassortments in influenza A viruses. J Bioinform Comput Biol 6(5):981–999. https://doi.org/10.1142/s0219720008003813
Tsuchiya E, Sugawara K, Hongo S, Matsuzaki Y, Muraki Y, Nakamura K (2002) Role of overlapping glycosylation sequons in antigenic properties, intracellular transport and biological activities of influenza A/H2N2 virus haemagglutinin. J Gen Virol 83(Pt 12):3067–3074. https://doi.org/10.1099/0022-1317-83-12-3067
Kaverin NV, Rudneva IA, Govorkova EA, Timofeeva TA, Shilov AA, Kochergin-Nikitsky KS, Krylov PS, Webster RG (2007) Epitope mapping of the hemagglutinin molecule of a highly pathogenic H5N1 influenza virus by using monoclonal antibodies. J Virol 81(23):12911–12917. https://doi.org/10.1128/JVI.01522-07
Iqbal M, Yaqub T, Reddy K, McCauley JW (2009) Novel genotypes of H9N2 influenza A viruses isolated from poultry in Pakistan containing NS genes similar to highly pathogenic H7N3 and H5N1 viruses. PLoS ONE 4(6):e5788. https://doi.org/10.1371/journal.pone.0005788
Matrosovich MN, Krauss S, Webster RG (2001) H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281(2):156–162. https://doi.org/10.1006/viro.2000.0799
Rashid S, Naeem K, Ahmed Z, Saddique N, Abbas MA, Malik SA (2009) Multiplex polymerase chain reaction for the detection and differentiation of avian influenza viruses and other poultry respiratory pathogens. Poult Sci 88(12):2526–2531. https://doi.org/10.3382/ps.2009-00262
Chen GW, Chang SC, Mok CK, Lo YL, Kung YN, Huang JH, Shih YH, Wang JY, Chiang C, Chen CJ, Shih SR (2006) Genomic signatures of human versus avian influenza A viruses. Emerg Infect Dis 12(9):1353–1360. https://doi.org/10.3201/eid1209.060276
Fan S, Hatta M, Kim JH, Halfmann P, Imai M, Macken CA, Le MQ, Nguyen T, Neumann G, Kawaoka Y (2014) Novel residues in avian influenza virus PB2 protein affect virulence in mammalian hosts. Nat Commun 5:5021. https://doi.org/10.1038/ncomms6021
Taft AS, Ozawa M, Fitch A, Depasse JV, Halfmann PJ, Hill-Batorski L, Hatta M, Friedrich TC, Lopes TJ, Maher EA, Ghedin E, Macken CA, Neumann G, Kawaoka Y (2015) Identification of mammalian-adapting mutations in the polymerase complex of an avian H5N1 influenza virus. Nat Commun 6:7491. https://doi.org/10.1038/ncomms8491
Rolling T, Koerner I, Zimmermann P, Holz K, Haller O, Staeheli P, Kochs G (2009) Adaptive mutations resulting in enhanced polymerase activity contribute to high virulence of influenza A virus in mice. J Virol 83(13):6673–6680. https://doi.org/10.1128/JVI.00212-09
Hulse-Post DJ, Franks J, Boyd K, Salomon R, Hoffmann E, Yen HL, Webby RJ, Walker D, Nguyen TD, Webster RG (2007) Molecular changes in the polymerase genes (PA and PB1) associated with high pathogenicity of H5N1 influenza virus in mallard ducks. J Virol 81(16):8515–8524. https://doi.org/10.1128/JVI.00435-07
Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J (2005) The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci U S A 102(51):18590–18595. https://doi.org/10.1073/pnas.0507415102.
Hatta M, Gao P, Halfmann P, Kawaoka Y (2001) Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293(5536):1840–1842. https://doi.org/10.1126/science.1062882
Song J, Xu J, Shi J, Li Y, Chen H (2015) Synergistic Effect of S224P and N383D Substitutions in the PA of H5N1 Avian Influenza Virus Contributes to Mammalian Adaptation. Sci Rep 5:10510. https://doi.org/10.1038/srep10510
Joseph U, Su YC, Vijaykrishna D, Smith GJ (2017) The ecology and adaptive evolution of influenza A interspecies transmission. Influenza Other Respir Viruses 11(1):74–84. https://doi.org/10.1111/irv.12412
Furuse Y, Suzuki A, Kamigaki T, Oshitani H (2009) Evolution of the M gene of the influenza A virus in different host species: large-scale sequence analysis. Virol J 6:67. https://doi.org/10.1186/1743-422X-6-67
Jie Y, Zheng H, Xiaolei L, Xinhua O, Dong Y, Yingchun S, Lingzhi L, Rengui Y (2018) Full-length genome analysis of an avian influenza A virus (H9N2) from a human infection in Changsha City. Future Virol 13(5):323–330. https://doi.org/10.2217/fvl-2017-0151
Shaw M, Cooper L, Xu X, Thompson W, Krauss S, Guan Y, Zhou N, Klimov A, Cox N, Webster R, Lim W, Shortridge K, Subbarao K (2002) Molecular changes associated with the transmission of avian influenza a H5N1 and H9N2 viruses to humans. J Med Virol 66(1):107–114. https://doi.org/10.1002/jmv.2118
Soubies SM, Volmer C, Croville G, Loupias J, Peralta B, Costes P, Lacroux C, Guérin JL, Volmer R (2010) Species-specific contribution of the four C-terminal amino acids of influenza A virus NS1 protein to virulence. J Virol 84(13):6733–6747. https://doi.org/10.1128/JVI.02427-09
Li S, Min JY, Krug RM, Sen GC (2006) Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349(1):13–21. https://doi.org/10.1016/j.virol.2006.01.005
Jiao P, Tian G, Li Y, Deng G, Jiang Y, Liu C, Liu W, Bu Z, Kawaoka Y, Chen H (2008) A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol 82(3):1146–1154. https://doi.org/10.1128/JVI.01698-07
Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Cox N (1998) Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279(5349):393–396
Wang J, Sun Y, Xu Q, Tan Y, Pu J, Yang H, Brown EG, Liu J (2012) Mouse-adapted H9N2 influenza A virus PB2 protein M147L and E627K mutations are critical for high virulence. PLoS ONE 7(7):e40752
Kong H, Ma S, Wang J, Gu C, Wang Z, Shi J, Deng G, Guan Y, Chen H (2019) Identification of key amino acids in the PB2 and M1 proteins of H7N9 influenza virus that affect its transmission in guinea pigs. J Virol 94(1):e01180–e01119
Lee MS, Deng MC, Lin YJ, Chang CY, Shieh HK, Shiau JZ, Huang CC (2007) Characterization of an H5N1 avian influenza virus from Taiwan. Vet Microbiol 124(3–4):193–201
Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P (2007) A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog 3(10):e141
Suguitan AL Jr, Matsuoka Y, Lau YF, Santos CP, Vogel L, Cheng LI, Orandle M, Subbarao K (2012) The multibasic cleavage site of the hemagglutinin of highly pathogenic A/Vietnam/1203/2004 (H5N1) avian influenza virus acts as a virulence factor in a host-specific manner in mammals. J Virol 86(5):2706–2714
Dankar SK, Wang S, Ping J, Forbes NE, Keleta L, Li Y, Brown EG (2011) Influenza A virus NS1 gene mutations F103L and M106I increase replication and virulence. Virol J 8:1–13
Subbarao K, Katz J (2000) Avian influenza viruses infecting humans. CMLS 57:1770–1784
Lycett SJ, Ward MJ, Lewis FI, Poon AFY, Kosakovsky Pond SL, Brown AL (2009) Detection of mammalian virulence determinants in highly pathogenic avian influenza H5N1 viruses: multivariate analysis of published data. J Virol 83(19):9901–9910
Ma S, Zhang B, Shi J, Yin X, Wang G, Cui P, Liu L, Deng G, Jiang Y, Li C, Chen H (2020) Amino acid mutations A286V and T437M in the nucleoprotein attenuate H7N9 viruses in mice. J Virol 94(2):e01530–e01519
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Sheela Ramamoorthy
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bedair, N.M., Sakr, M.A., Mourad, A. et al. Molecular characterization of the whole genome of H9N2 avian influenza virus isolated from Egyptian poultry farms. Arch Virol 169, 99 (2024). https://doi.org/10.1007/s00705-024-06018-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-024-06018-2