Abstract
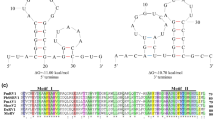
Paris mitovirus 1 (ParMV1) is a positive-sense RNA virus that was detected in diseased Paris polyphylla var. yunnanensis plants in Wenshan, Yunnan. The complete genome sequence of ParMV1 is 2,751 nucleotides in length, and the genome structure is typical of mitoviruses. The ParMV1 genome has a single open reading frame (ORF; nt 358-2,637) that encodes an RNA-dependent RNA polymerase (RdRp) with a predicted molecular mass of 86.42 kDa. ParMV1 contains six conserved motifs (Ι-VΙ) that are unique to mitoviruses. The 5′ and 3′ termini of the genome are predicted to have a stable secondary structure, with the reverse complementary sequence forming a panhandle structure. Comparative genome analysis revealed that the RdRp of ParMV1 shares 23.1-40.6% amino acid (aa) and 32.3-45.7% nucleotide (nt) sequence identity with those of other mitoviruses. Phylogenetic analysis based on RdRp aa sequences showed that ParMV1 clusters with mitoviruses and hence should be considered a new member of the genus Mitovirus in the family Mitoviridae. This is the first report of a novel mitovirus infecting Paris polyphylla var. yunnanensis.


Similar content being viewed by others
Data availability
The dataset generated during the current study is available in the GenBank database (accession no. MT269666).
References
Qin XJ, Ni W, Chen CX et al (2018) Seeing the light: shifting from wild rhizomes to extraction of active ingredients from above-ground parts of Paris polyphylla var. yunnanensis. J Ethnopharmacol 224:134–139
Zhou N, Xu L, Park SM et al (2021) Genetic diversity, chemical components, and property of biomass Paris polyphylla var. yunnanensis. Front Bioeng Biotechnol 9:713860
Lan P, Zhao J, Zhou Y et al (2018) Complete genome sequence of Paris mosaic necrosis virus, a distinct member of the genus Potyvirus. Arch Virol 163:787
Dong JH, Ding M, Fang Q et al (2007) Molecular identifcation of a Potexvirus isolate infecting Pairs polyphylla var. Yunnanensis and analysis of 3’terminal sequence. Acta Phytopathol Sin 37:237–241
Wen GS, Yang LY, Anane RF et al (2019) First report of Pepper mild mottle virus in Pairs polyphylla var. Yunnanensis in China. Plant Dis 103:3289
Chen L, Anane RF, Wang Z et al (2021) Whole-genome sequence analysis of paris virus 1: a novel member of the genus Potyvirus infecting Paris polyphylla var. yunnanensis. Arch Virol 165:985–988
Chen ZL, Anane RF, Wang Z et al (2021) Complete genome sequence analysis of a novel coguvirus isolated from Paris polyphylla var. yunnanensis. Arch Virol 166:2045–2050
Chen L, Anane RF, Wang Z et al (2021) Characterization of a novel Tombusviridae species isolated from Paris polyphylla var. yunnanensis. Arch Virol. https://doi.org/10.1007/s00705-021-05191-y
Chen Y, Shang H, Yang H et al (2017) A mitovirus isolated from the phytopathogenic fungus Alternaria brassicicola. Arch Virol 162:2869–2874
Zhang T, Li W, Chen H, Yu H (2015) Full genome sequence of a putative novel mitovirus isolated from Rhizoctonia cerealis. Arch Virol 160:1815–1818
Hillman BI, Cai G (2013) Chapter six-the family Narnaviridae: simplest of RNA viruses. Adv Virus Res 86:149–176
Deng F, Boland GJ (2006) Attenuation of virulence in Sclerotinia homoeocarpa during storage is associated with latent infection by Ophiostoma mitovirus 3a. Eur J Plant Pathol 114:127–137
Sahin E, Akata I (2019) Complete genome sequence of a novel mitovirus from the ectomycorrhizal fungus Geopora sumneriana. Arch Virol 164:2853–2857
Cole TE, Hong Y, Brasier CM et al (2000) Detection of an RNA-dependent RNA polymerase in mitochondria from a mitovirus-infected isolate of the Dutch Elm disease fungus, Ophiostoma novo-ulmi. Virology 268:239–243
Hong Y, Cole TE, Brasier CM et al (1998) Evolutionary relationships among putative RNA-dependent RNA polymerases encoded by a mitochondrial virus-like RNA in the Dutch elm disease fungus, Ophiostoma novo-ulmi, by other viruses and virus-like RNAs and by the Arabidopsis mitochondrial genome. Virology 246:158–169
Hong Y, Dover SL, Cole TE et al (1999) Multiple mitochondrial viruses in an isolate of the Dutch Elm disease fungus Ophiostoma novo-ulmi. Virology 258:118–127
Polashock JJ, Hillman BI (1994) A small mitochondrial double-stranded (ds) RNA element associated with a hypovirulent strain of the chestnut blight fungus and ancestrally related to yeast cytoplasmic T and W dsRNAs. Proc Natl Acad Sci U S A 91:8680–8684
Rogers HJ, Buck KW, Brasier CM (1987) A mitochondrial target for the double-stranded RNAs in diseased isolates of the fungus that causes dutch elm disease. Nature 329:558–560
Zheng L, Zhao J, Liang X et al (2019) Complete genome sequence of a novel mitovirus from the wheat stripe rust fungus Puccinia striiformis. Arch Virol 164:897–901
Ran H, Liu L, Li B et al (2016) Co-infection of a hypovirulent isolate of Sclerotinia sclerotiorum with a new botybirnavirus and a strain of a mitovirus. Virol J 13:92
Khalifa ME, Pearson MN (2013) Molecular characterization of three mitoviruses co-infecting a hypovirulent isolate of Sclerotinia sclerotiorum fungus. Virology 441:22–30
Stielow B, Klenk HP, Winter S et al (2011) A novel Tuber aestivum (Vittad.) mitovirus. Arch Virol 156:1107–1110
Giovannetti M, Azzolini D, Citernesi AS (1999) Anastomosis formation and nuclear and protoplasmic exchange in arbuscular mycorrhizal fungi. Appl Environ Microbiol 65:5571–5575
Polashock JJ, Bedker PJ, Hillman BI (1997) Movement of a small mitochondrial double-stranded RNA element of Cryphonectria parasitica: ascospore inheritance and implications for mitochondrial recombination. Mol Gen Genet 256:566–571
Nerva L, Vigani G, Silvestre DD et al (2019) Biological and molecular characterization of Chenopodium quinoa mitovirus 1 reveals a distinct small RNA response compared to those of cytoplasmic RNA Viruses. J Virol 93:01998–02015
Nibert ML, Vong M, Fugate KK et al (2018) Evidence for contemporary plant mitoviruses. Virology 518:14–24
Bruenn JA (1991) Relationships among the positive-strand and double-strand RNA viruses as viewed through their RNA-dependent RNA polymerases. Nucleic Acids Res 19:217–226
Bruenn JA (1993) A closely related group of RNA-dependent RNA polymerases from double-stranded RNA viruses. Nucleic Acids Res 21:5667–5669
Habili N, Symons RH (1989) Evolutionary relationships between luteoviruses and other RNA plant viruses based on sequence motifs in their putative RNA polymerases and nucleic acid helicases. Nucleic Acids Res 17:9543–9555
Koonin EV (1991) The phylogeny of RNA-dependent RNA poly-merases of positive-strand RNA viruses. J Gen Virol 72:2197–2206
Nibert ML (2017) Mitovirus UGA(Trp) codon usage parallels that of host mitochondria. Virology 507:96
Jukes TH, Osawa S (1990) The genetic code in mitochondria and chloroplasts. Experientia 46:1117–1126
Xu Z, Wu S, Liu L, Cheng J et al (2015) A mitovirus related to plant mitochondrial gene confers hypovirulence on the phytopathogenic fungus Sclerotinia sclerotiorum. Virus Res 197:127–136
Doherty M, Coutts RHA, Brasier CM et al (2006) Sequence of RNA-dependent RNA Polymerase genes provides evidence for three more distinct mitoviruses in Ophiostoma Novo-ulmi Isolate Ld. Virus Genes 33:41–44
Xie J, Wei D, Jiang D et al (2006) Characterization of debilitation-associated mycovirus infecting the plant-pathogenic fungus Sclerotinia sclerotiorum. J Gen Virol 87:241–249
Funding
This study was funded by the National Natural Science Foundation of China (81860774).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Stephen John Wylie.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, Z., Chen, L., Anane, R.F. et al. Complete genome sequence of a novel mitovirus detected in Paris polyphylla var. yunnanensis. Arch Virol 167, 645–650 (2022). https://doi.org/10.1007/s00705-021-05339-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05339-w