Abstract
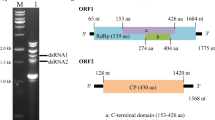
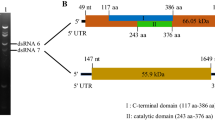
Mycoviruses from Metarhizium anisopliae have been extensively studied, but their sequences have yet to be deposited in the NCBI database. In the present study, we characterized a new partitivirus obtained from the entomogenous fungus Metarhizium brunneum, named “Metarhizium brunneum partitivirus 1” (MbPV1). The complete genome of MbPV1, determined by metagenomic sequencing, RT-PCR, and RACE, comprised two dsRNA segments of 1,829 bp and 1,720 bp, respectively. Both dsRNAs contained a single open reading frame (ORF), encoding a putative RNA-dependent RNA polymerase (RdRp) and a coat protein (CP), respectively. The sequences of the RdRp and CP showed the highest similarity (61.4% and 44.4% identity, respectively) to those of Colletotrichum eremochloae partitivirus 1 (CePV1), which were obtained from the NCBI database. A phylogenetic tree based on the RdRp sequence showed that MbPV1 clustered with members of the proposed genus “Epsilonpartitivirus”, belonging to family Partitiviridae. Here, we propose that MbPV1 is a member of a new species of the proposed genus “Epsilonpartitivirus”. This is the first sequence data report of a new mycovirus from a member of the genus Metarhizium.




Similar content being viewed by others
References
Ghabrial SA, Castón JR, Jiang D, Nibert ML, Suzuki N (2015) 50-plus years of fungal viruses. Virology 479–480:356–368
Santos V, Mascarin GM, da Silva Lopes M, Alves MCDF, Rezende JM, Gatti MSV, Dunlap CA, Delalibera Júnior Í (2017) Identification of double-stranded RNA viruses in Brazilian strains of Metarhizium anisopliae and their effects on fungal biology and virulence. Plant Gene 11:49–58
de la Paz Giménez-Pecci M, Bogo MR, Santi L, Moraes CKd, Corrêa CT, Henning Vainstein M, Schrank A (2002) Characterization of mycoviruses and analyses of chitinase secretion in the biocontrol fungus Metarhizium anisopliae. Curr Microbiol 45:334–339
Quinelato S, Golo PS, Perinotto WMS, Sá FA, Camargo MG, Angelo IC, Moraes AML, Bittencourt VREP (2012) Virulence potential of Metarhizium anisopliae s.l. isolates on Rhipicephalus (Boophilus) microplus larvae. Vet Parasitol 190:556–565
Perinotto WMS, Golo PS, Coutinho Rodrigues CJB, Sá FA, Santi L, Beys da Silva WO, Junges A, Vainstein MH, Schrank A, Salles CMC, Bittencourt VREP (2014) Enzymatic activities and effects of mycovirus infection on the virulence of Metarhizium anisopliae in Rhipicephalus microplus. Vet Parasitol 203:189–196
Vainio EJ, Chiba S, Ghabrial SA, Maiss E, Roossinck M, Sabanadzovic S, Suzuki N, Xie JT, Nibert M, Consortium IR (2018) ICTV virus taxonomy profile: Partitiviridae. J Gener Virol 99:17–18
Nerva L, Silvestri A, Ciuffo M, Palmano S, Varese GC, Turina M (2017) Transmission of Penicillium aurantiogriseum partiti-like virus 1 to a new fungal host (Cryphonectria parasitica) confers higher resistance to salinity and reveals adaptive genomic changes. Environ Microbiol 19:4480–4492
Gilbert KB, Holcomb EE, Allscheid RL, Carrington JC (2019) Hiding in plain sight: New virus genomes discovered via a systematic analysis of fungal public transcriptomes. PLoS One 14:e0219207
Herrero N, Dueñas E, Quesada-Moraga E, Zabalgogeazcoa I (2012) Prevalence and diversity of viruses in the entomopathogenic fungus Beauveria bassiana. Appl Environ Microb 78:8523–8530
Katoh K, Rozewicki J, Yamada KD (2017) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 4:1–7
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
te Velthuis AJW (2014) Common and unique features of viral RNA-dependent polymerases. Cell Mol Life Sci 71:4403–4420
Bruenn JA (2003) A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res 31:1821–1829
Bruenn JA (1991) Relationships among the positive strand and double-strand RNA viruses as viewed through their RNA-dependent RNA polymerases. Nucleic Acids Res 19:217–226
Shi N, Yang G, Wang P, Wang Y, Yu D, Huang B (2019) Complete genome sequence of a novel partitivirus from the entomogenous fungus Beauveria bassiana in China. Arch Virol 164:3141–3144
Funding
This work was supported by the National Natural Science Foundation of China (Grant nos. 31772226, 31471821 and 30770008).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Robert H.A. Coutts.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, P., Yang, G., Shi, N. et al. Molecular characterization of a new partitivirus, MbPV1, isolated from the entomopathogenic fungus Metarhizium brunneum in China. Arch Virol 165, 765–769 (2020). https://doi.org/10.1007/s00705-019-04517-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04517-1