Abstract
Ectomycorrhizal (EM) associations can promote the dominance of tree species in otherwise diverse tropical forests. These EM associations between trees and their fungal mutualists have important consequences for soil organic matter cycling, yet the influence of these EM-associated effects on surrounding microbial communities is not well known, particularly in neotropical forests. We examined fungal and prokaryotic community composition in surface soil samples from mixed arbuscular mycorrhizal (AM) and ectomycorrhizal (EM) stands as well as stands dominated by EM-associated Oreomunnea mexicana (Juglandaceae) in four watersheds differing in soil fertility in the Fortuna Forest Reserve, Panama. We hypothesized that EM-dominated stands would support distinct microbial community assemblages relative to the mixed AM-EM stands due to differences in carbon and nitrogen cycling associated with the dominance of EM trees. We expected that this microbiome selection in EM-dominated stands would lead to lower overall microbial community diversity and turnover, with tighter correspondence between general fungal and prokaryotic communities. We measured fungal and prokaryotic community composition via high-throughput Illumina sequencing of the ITS2 (fungi) and 16S rRNA (prokaryotic) gene regions. We analyzed differences in alpha and beta diversity between forest stands associated with different mycorrhizal types, as well as the relative abundance of fungal functional groups and various microbial taxa. We found that fungal and prokaryotic community composition differed based on stand mycorrhizal type. There was lower prokaryotic diversity and lower relative abundance of fungal saprotrophs and pathogens in EM-dominated than AM-EM mixed stands. However, contrary to our prediction, there was lower homogeneity for fungal communities in EM-dominated stands compared to mixed AM-EM stands. Overall, we demonstrate that EM-dominated tropical forest stands have distinct soil microbiomes relative to surrounding diverse forests, suggesting that EM fungi may filter microbial functional groups in ways that could potentially influence plant performance or ecosystem function.
Similar content being viewed by others
Introduction
Plants often influence surrounding soils via interactions with microbial organisms (Zak et al. 2003). Predominant among these plant–microbe interactions are mycorrhizal associations between fungi and plant roots (Hawkes et al. 2007). Two main types of mycorrhizal association, namely, arbuscular mycorrhizal (AM) and ectomycorrhizal (EM), can impact plant health (Revillini et al. 2016), litter decomposition (Jacobs et al. 2018), nutrient cycling (Phillips et al. 2013), and soil organic matter dynamics (Frey 2019). Importantly, mycorrhizal fungi facilitate plant acquisition of soil nutrients such as nitrogen and phosphorous to support plant growth and nutrition (Smith and Smith 2011). However, bacteria, archaea, and non-mycorrhizal fungi also contribute to mycorrhizal plant and ecosystem effects as they can influence nutrient transformation processes, promote plant root exudation, and benefit plant growth (Tarkka et al. 2018; Sangwan and Prasanna 2022; Berrios et al. 2023). These non-mycorrhizal microbial communities are likely responsible for the extended plant-soil effects of mycorrhizal associations that can influence overall forest population dynamics in temperate ecosystems (Bennett et al. 2017). However, we know very little about how these “mycorrhizosphere” (Rambelli 1973) microbiomes manifest in tropical ecosystems that have differing patterns of productivity, species diversity, and soil functionality (Barlow et al. 2018).
Neotropical forests are generally a matrix of primarily AM-associating tree species interspersed with fewer EM-associated species (McGuire et al. 2012), but EM-associated species can also form stands where the majority of the total basal area comprises a single species (Hart et al. 1989; Peh et al. 2011). These EM-dominated stands often differ significantly from surrounding mixed-mycorrhizal forest stands in important ecosystem characteristics (Torti et al. 2001), with slower decomposition rates (McGuire et al. 2010) and lower soil inorganic nutrient availability (Corrales et al. 2016b) than adjacent mixed AM-EM forest stands. Ectomycorrhizal-dominated stands in these forests are also associated with changes to the relative abundance of some fungal functional guilds (Seyfried et al. 2022), but the impact of EM dominance on the broader complex soil microbiome, particularly prokaryotic communities, is still unclear due to a relative lack of data from neotropical forests.
Mycorrhizal fungi often interact with surrounding microbial communities in ways that create favorable ecological conditions for their plant partners (Uroz et al. 2019). These interactions may play an important role in the formation or proliferation of EM-dominated forest stands. For example, EM fungi are thought to outcompete saprotrophic organisms for nutrients in soil organic matter (SOM; Averill and Hawkes 2016) due to their ability to produce SOM-degrading extracellular enzymes (Pellitier and Zak 2018). This fungal interguild competition can increase soil carbon (C)-to-nutrient ratios, slowing C and inorganic nutrient cycling (Fernandez and Kennedy 2016). These changes could decrease overall microbial C and nutrient availability, potentially resulting in lower overall microbial diversity with increasing EM dominance (Eagar et al. 2021; Heděnec et al. 2023), as well as downstream impacts on copiotrophic or oligotrophic soil microbiota (Nemergut et al. 2010). Ectomycorrhizal-associated changes to non-mycorrhizal microbial communities associated with soil organic matter cycling could potentially further promote positive feedbacks to slow SOM cycling and create a competitive advantage for EM mutualisms. Arbuscular mycorrhizal fungi are often thought to scavenge inorganic nutrients from soil and must rely on surrounding microbial communities to degrade SOM (van Der Heijden et al. 2015). Mycorrhizal fungi also influence the activity and abundance of soil fungal pathogens (Borowicz 2001; Veresoglou and Rillig 2012). Both AM and EM fungi can confer pathogen resistance to their plant hosts through the release of volatile organic compounds (Dreischhoff et al. 2020) or extracellular secretion of secondary metabolites (Pellegrin et al. 2015). However, EM relationships can generate greater conspecific benefits for pathogen suppression than AM, potentially promoting EM dominance (Liang et al. 2020) and resulting in greater conmycorrhizal plant recruitment (Delavaux et al. 2023). Overall, the effects of mycorrhizal associations on surrounding microbial community function are highly context-dependent, with their outcome varying based on mycorrhizal fungal species (Emmett et al. 2021), plant species and litter quality (Fernandez et al. 2019), climate (Bennett and Classen 2020), and soil parent material (Seyfried et al. 2021b). Quantifying the effects of EM dominance on the different constituents of the soil microbiome could provide valuable insight to help contextualize the effects of mycorrhizal associations on surrounding soil microbiomes.
To characterize soil microbiome responses to EM tree dominance in a neotropical forest, we conducted a study comparing bulk soil fungal and prokaryotic communities between EM-dominated (by Oreomunnea mexicana at > 50% basal area per stand) and mixed AM-EM stands of highly diverse lower montane tropical forests in western Panama. While these sites also contain ~ 14 other EM tree species, those species occur in low abundance, and O. mexicana is the only one to form monodominant stands (Prada et al. 2017). Variation in parent material and geology among these stands leads to the formation of soils differing in nutrient availability, pH, and base saturation (Prada et al. 2017; Seyfried et al. 2021a), allowing for the investigation of microbial relationships with stand mycorrhizal type across a range of soil fertilities. We hypothesized that EM-dominated stands would be associated with distinct fungal and prokaryotic community assemblages relative to the AM-EM mixed stands. Given that EM dominance can slow SOM cycling and reduce nutrient availability, we present three predictions related to microbiome differences between stand mycorrhizal types (1) that EM stands would be associated with decreased microbial community diversity and heterogeneity compared to mixed AM-EM stands; (2) that EM stands would be associated with decreased relative abundance of saprotrophic and pathogenic fungal functional guilds, as well as bacteria and archaea associated with SOM cycling, compared to AM-EM mixed stands; and (3) that microbial communities in EM-dominated stands would have tighter Procrustean correspondence between general fungal and prokaryotic communities, as EM fungi could be acting to filter the soil microbiome to select microbial organisms that may contribute to an EM competitive advantage.
Methods
Study design
This study was conducted in four adjacent watersheds in the Fortuna Forest Reserve of western Panama: Alto Frio, Honda, Hornito, and Pinola/Zorro. We chose to use Pinola/Zorro as a paired site (collectively referred to hereafter as “Pinola”) because there were no EM-dominated stands at the Pinola site and the Zorro site was the closest alternative (~ 800 m away, although on differing parent material). These diverse neotropical montane forests differ in elevation, parent material, soil chemistry, and tree species diversity, containing between 61 and 153 (primarily AM-associating) species/ha (Prada et al. 2017), but also large (> 1 ha) patches where > 50% of the basal area is made up of the EM-associated species O. mexicana (Table 1; Dalling and Turner 2021). In April 2015, three soil samples were collected from one EM-dominated and from one mixed AM-EM stand within each watershed (4 watersheds × 2 stand mycorrhizal stand types × 3 replicates = 24 samples in total). The soil samples were collected from 0–10 cm depth using a 10 cm diameter soil corer after removing the leaf litter layer; thus, ~ 785 cm3 of soil was collected for each sample. Samples contained soil from both organic and mineral horizons, as we did not separate these layers due to high variability in organic horizon depth. Within each stand, the three soil samples collected in each stand were from locations separated from one another by ~ 5 m, and all samples were collected > 1 m away from the nearest tree trunk to limit localized effects of individual species on microbial community composition. Soil samples were stored in insulated coolers with ice packs and transported to the Smithsonian Tropical Research Institute’s Naos Marine and Molecular Laboratories in Panama within 2 days of collecting. At Naos, samples were passed through a 2-mm sieve (sterilized with 70% EtOH between samples), and roots/course organic matter was removed. After processing, samples were stored at − 80 °C for up to 1 week prior to DNA extraction.
DNA extraction and sequencing
To characterize the fungal and prokaryotic communities, DNA was extracted from 0.25 g of soil using the PowerSoil DNA Isolation Kit (MO BIO Laboratories, Carlsbad, USA). DNA extracts were submitted to the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign for PCR amplification using a Fluidigm Access Array IFC chip (see: Cronn et al. 2012), which allows for amplification of multiple primer sets simultaneously (Fluidigm, San Francisco, USA), and Illumina sequencing (Illumina, San Diego, USA). Specific details for the PCR procedures and mixtures as well as library generation can be found in Suriyavirun et al. (2019). Fungal communities were assessed via the ITS2 gene region using ITS3 and ITS4 (White et al. 1990) primers to amplify DNA. We assessed bacterial and archaeal communities via the bacterial and archaeal 16S rRNA genes. The samples were amplified using V4_515f forward (Parada et al. 2016) and V4_806r reverse (Apprill et al. 2015) primers. Sequence information for all primers can be found in Table S1. Both ITS and 16S amplicons were sequenced via Illumina HiSeq bulk 2 × 250 bp V2. Sequence data are publicly available from the NCBI SRA database under accession number PRJNA1027860.
To assess variation in microbial communities, we generated amplicon sequence variants (ASVs) from our sequence data using the DADA2 bioinformatics pipeline (Version 1.16; Callahan et al. 2016). Sequences were quality filtered and denoised, forward and reverse reads were merged, and chimeric sequences were removed using recommended parameters for 16S and ITS genes (Callahan et al. 2020). Taxonomy was assigned via the naïve Bayesian DADA2 classifier (Wang et al. 2007), using the SILVA R138.1 (Quast et al. 2012) and UNITE V9.0 (Nilsson et al. 2019) reference databases for 16S and ITS taxonomic assignments, respectively. Final count numbers were relativized without rarefaction (McMurdie and Holmes 2014) via a Hellinger transformation prior to statistical analysis. Final datasets consisted of 1043 fungal, 25,817 bacterial, and 96 archaeal ASVs. More information about sequencing depth and quality can be found in supplementary Table S2 and supplementary Fig. S1. Due to being derived from the same primer sets, bacterial and archaeal communities were analyzed together. We assigned fungi to saprotrophic, pathogenic, and EM functional guilds using the FUNGuild database (Nguyen et al. 2016), classifying assignments to guilds with “probable” or “highly probable” confidence scores.
Statistical analysis
Statistical analyses were performed in the R statistical environment (Version 4.3.2; R Core Team 2013). To understand how stand mycorrhizal type influenced microbial alpha diversity (i.e., relative ASV richness and diversity), we used Hill numbers for orders of q (q = 0, 1, 2) using the hillR package (Version 0.5.2; Li 2018). Hill numbers provide the “effective number of species” or “species equivalents” (MacArthur 1965; Hill 1973; Jost 2007; Chao et al. 2014): q = 0 is representative of richness, where all species are weighted equally; q = 1 is the exponential of Shannon entropy representative of diversity, where species are weighted by their proportional abundance; and q = 2 is equivalent to the inverse of Simpson’s index, where rare species are down-weighted. To assess differences in beta diversity patterns between stand mycorrhizal types, we used distance-based redundancy analyses (dbRDA) in the VEGAN package (Version 2.6–4; Oksanen et al. 2010) on a Jaccard distance matrix of Hellinger transformed fungal and prokaryotic ASV abundances. Stand mycorrhizal type was a fixed factor in the dbRDA, and the sampling site was partialed out via the Condition() arguments in the dbrda() function in VEGAN. We also depict these relationships visually through the dbRDA ordination without fixed or partialed effects. To assess the relative variation explained by site and stand mycorrhizal type, we used variance partitioning via the varpart() function in VEGAN on the Jaccard distance matrix for both fungal and bacterial/archaeal communities. To understand the correspondence between fungal and bacterial/archaeal communities, we calculated the procrustean association metric (PAM) based on the output of the residuals() function from procrustean correspondence between fungal and bacterial/archaeal dbRDA ordinations, protest() function in VEGAN. We used linear mixed models to test the effects of stand mycorrhizal type on fungal and prokaryotic Hill numbers, the relative abundance/richness of fungal functional guilds (saprotrophs, pathogens, and EM fungi), and PAM, with stand mycorrhizal type as a fixed effect and site location as a random effect. Finally, to understand whether community dissimilarity between fungal and bacteria/archaeal communities was related to the relative abundance/richness of EM, saprotrophic, or pathogenic fungi, we used a general linear model to examine the relationship between PAM and the relative abundance of fungal functional groups. We used DESeq2 (Version 3.18; Love et al. 2014) to determine bacterial, archaeal, and fungal phyla, classes, orders, families, and genera that differed significantly in abundance between stand mycorrhizal types, as this approach accounts for multiple comparisons and overdispersion among taxonomic count numbers. Significance for all statistical test was assessed as p < 0.05.
Results
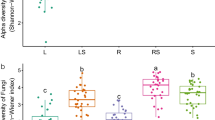
Ectomycorrhizal-dominated stands had significantly lower alpha diversity (Hill numbers) for bacterial/archaeal communities at all orders of q, with 27% fewer effective number of species at q = 0, 40% fewer effective number of species at q = 1, and 46% fewer effective number of species at q = 2 (p < 0.05, Fig. 1). In contrast, we found no significant difference between EM-dominated and mixed AM-EM stands for fungal Hill numbers at any order of q (Fig. 1).
Mean (25th, 75th percentile, lowest and highest values observed) fungal and bacterial alpha diversity between ectomycorrhizal (EM) and mixed arbuscular mycorrhizal (AM)-EM stand types using Hill numbers depicting effective number of species with all ASVs weighted equally (q = 0), ASVs weighted based on their proportional abundance (q = 1), and rare ASVs down-weighted (q = 2)
The relative abundances and ASV richness of fungal functional guilds differed significantly between stand mycorrhizal types. The relative abundance of EM fungal ASVs was significantly greater in EM-dominated stands (average 180%) relative to mixed AM-EM stands (F1,19 = 11.75, p = 0.002; Fig. 2). Richness of EM fungal ASVs was also significantly greater (average 170%) relative to mixed AM-EM stands (F1,19 = 59.7, p < 0.001; Fig. 2). On average, saprotrophic fungal ASV relative abundance was 56% lower in EM-dominated than mixed AM-EM stands (F1,19 = 6.33, p = 0.02), with ASV richness of fungal saprotrophs decreasing 27.5% from EM-dominated to AM-EM mixed stands (F1,19 = 5.35, p = 0.03). Pathogenic fungal ASV relative abundance was 50% lower in EM-dominated stands than AM-EM mixed stands (F1,19 = 10.2, p = 0.004; Fig. 2); however, pathogenic ASV richness was not different between stand mycorrhizal types. We did not detect significant correlations among the ASV relative abundances of saprotrophic, pathogenic, and EM functional guilds (p > 0.05). We also did not find a significant difference between stand mycorrhizal types in the relative abundance of ASVs not assigned to saprotrophic, pathogenic, or EM functional guilds (average 68.2%, p > 0.05; Fig. S2).
Fungal and bacterial/archaeal communities both differed significantly between stand mycorrhizal types, but the ways in which these communities differed, and the potential drivers of these differences were not the same for fungi and prokaryotes. Stand mycorrhizal type explained 7.71% of the variation in fungal community composition among samples (dbRDA: F1,19 = 2.81, p = 0.001), while site explained 13.1% of variations. Fungal communities in EM-dominated stands had significantly greater dispersion than those in mixed AM-EM stands (betadisper: F1,21 = 5.28, p = 0.03; Fig. 3a). Stand mycorrhizal type explained 15.3% of the variation in bacterial/archaeal community composition (F1,19 = 5.02, p = 0.001), while site explained 14.1% of the variation. We did not detect significant stand mycorrhizal type differences in bacterial/archaeal community homogeneity (Fig. 3b).
Fungal (a) and bacterial (b) community ordinations with axes indicating the proportion of community variation explained by the first and second multidimensional dimension scaling (MDS). Procrustean residuals between fungal and bacterial community ordinations with site-specific differences shown in the panel and overall difference between stand mycorrhizal type shown in insert (c). Relationship between procrustean residuals and fungal pathogen relative abundance (d)
There was a significant Procrustean rotational similarity correlation between fungal and bacterial/archaeal ordinations (R2 = 0.78, p = 0.001). However, Procrustean residuals (PAM) were significantly lower (i.e., greater resemblance) in EM-dominated than mixed AM-EM stands (Fig. 3c). Further, there was a significant positive relationship between PAM and fungal pathogen relative abundance (R2 = 0.16, p = 0.03; Fig. 3d). We did not detect a similar relationship between PAM and the relative abundance of saprotrophic or EM fungi (Fig. S3; p > 0.05).
The dominance of EM-associated trees was significantly correlated with the abundance of fungal and bacterial/archaeal groups at various phylogenetic levels, with some taxonomic groups exhibiting increased and others decreased relative abundances in mixed AM-EM compared to EM-dominated stands. We found that 3 fungal phyla (Mortierellomycota, Kickxellomycota, and Chytridiomycota) were less abundant in EM-dominated than mixed AM-EM stands (Table S3). For fungi overall, 5 classes, 6 orders, 12 families, and 18 genera were less abundant in EM-dominated stands, while three classes, 4 orders, 8 families, and 9 genera were more abundant in these stands (Table S3). For prokaryotes, 10 among the 16 phyla that responded to stand mycorrhizal type were less abundant in EM-dominated stands (e.g., Bacteroidota, Myxococcota, Firmicutes, Gemmatimonadta, Methylomirabilota, Latescibacterota, Nitrospirota, MBNT15, Entotheonellaeota, and SAR324), while Proteobacteria, Acidobacteriota, Planctomyceteota, RCP2-54, WPS-2, and Armatimonadota were more abundant in EM-dominated stands (Table S4, Fig. 4). For prokaryotes overall, 29 classes, 86 orders, 153 families, and 227 genera were less abundant in EM-dominated stands (Table S4). Comparatively, far fewer prokaryotic groups were more abundant in EM-dominated stands, with 8 classes, 19 orders, 20 families, and 32 genera falling into this category (Table S4). However, these groups often represented the most abundant bacterial/archaeal groups overall, as shown in the graphical depictions of the relative abundance of the top 10 bacterial groups at these phylogenetic levels (Figs. S4–S7).
Relative abundance of the top 10 fungal and bacterial/archaeal phyla in ectomycorrhizal (EM-dominated) and arbuscular mycorrhizal-EM (AM-EM mixed) tree stands. Adjacent to phylum names, * indicates a group significantly more abundant in AM-EM mixed plots while + indicates a group significantly more abundant in EM-dominated plots
Discussion
We found a strong relationship between EM dominance and bulk soil microbiome composition in diverse neotropical montane forests. The relative abundance of EM-associating tree species in tropical forests is associated with significant shifts in soil properties (Barceló et al. 2022), yet it remains unclear how these shifts influence soil microbial composition. In support of our hypothesis, we found clear differentiation in soil microbial communities between stand mycorrhizal types across a range of forests differing in parent material and soil chemical properties, with stand mycorrhizal type explaining slightly greater variation in prokaryotic communities than site location. In relation to our predictions, (1) we demonstrate that EM-dominated forest stands maintain a less diverse prokaryotic microbiome than mixed AM-EM stands, (2) EM-dominated stands have lower relative abundance of fungal saprotrophs and pathogens than surrounding AM-EM mixed stands with taxonomic shifts of prokaryotes aligning with expected functional shifts for SOM cycling, and (3) correspondence between fungal and prokaryotic communities was greater in EM-dominated stands than in AM-EM mixed stands, but this did not correlate with the relative abundance of EM fungi. Our findings suggest that mycorrhizal fungi may affect the composition of non-mycorrhizal communities either directly or indirectly, through mycorrhizal effects on plant communities (Liang et al. 2020) and soil nutrient economies (Corrales et al. 2016b).
Ectomycorrhizal tree and fungal microbiome recruitment may have influenced the lower prokaryotic diversity we observed in EM-dominated than in mixed AM-EM stands. Tree species can independently recruit distinct bacterial communities (Oh et al. 2012). The EM-dominated stands we studied have a much lower overall tree species diversity than surrounding AM-EM mixed stands (Prada et al. 2017), potentially explaining higher prokaryotic community diversity in these stands. At our study site, EM leaf litter is not necessarily lower quality than AM leaf litter (Seyfried et al. 2021b). However, leaf litter in EM-dominated stands may be more chemically homogenous than leaf litter in mixed species AM stands where a high diversity of AM tree species contributes litter ranging widely in chemical quality to the forest floor (Seyfried et al. 2021b). Chemical heterogeneity could increase overall niche breath for microorganisms to support a highly diverse bacterial/archaeal community in AM-EM mixed stands. Additionally, mycorrhizal fungi may affect prokaryote communities directly by recruiting specific hyphosphere bacterial communities (Liu et al. 2018; Heděnec et al. 2020; Zhang et al. 2022). Specifically, AM fungi can support bacterial growth to facilitate inorganic nutrient transformations (Wang et al. 2023), while EM fungi may compete with free-living decomposers for organic nutrients (Fernandez and Kennedy 2016). These mycorrhizal hyphosphere responses also may be driven by differing patterns of belowground C allocation and root/fungal exudation between AM- and EM-associating tree and fungal species (Xu et al. 2023). Mycorrhizae and prokaryotic communities show strong patterns of interactions that may be driven by above- and belowground functional differences between AM and EM guilds.
Shifts in alpha diversity in response to stand mycorrhizal type also corresponded to functional changes among prokaryotic taxa. The influence of EM-dominated versus AM-EM mixed stand type on microbiome recruitment could have driven the lower relative abundance of bacterial/archaeal groups associated with inorganic nutrient transformation processes, such as Nitrospirota (Myrold 2021), in EM-dominated stands relative to AM-EM mixed stands. Microbiota beneficial to AM fungi could have been inhibited either directly, or indirectly through EM effects on inorganic N availability (Phillips et al. 2013; Corrales et al. 2016b). Further, there may be a relationship between these changes to functionally important taxa and EM-associated C dynamics. Ectomycorrhizal-dominated stands favored bacterial groups associated with diminished C mineralization activities, with greater Acidobacteriota and lower Bacteroidota relative abundance (Fierer et al. 2007) than in mixed AM-EM stands. These differences in the taxonomic composition of prokaryotic communities suggest that there may also be lower overall rates of C cycling and SOM decomposition in AM-EM mixed stands than in EM-dominated stands. Alternatively, O. mexicana may produce allelochemicals, as has been observed for other members of Juglandaceae (Jose and Gillespie 1998), which could be responsible for directly inhibiting specific soil microbiota (Revillini et al. 2023). This mechanism could potentially explain negative plant-soil feedbacks associated with O. mexicana legacy, although further investigation is necessary to determine the presence and identity of any possible allelopathic chemicals this plant could produce. Overall, suppression of C and N cycling by EM fungi and low tree diversity resulting in chemically homogenous root and leaf litter inputs in EM-dominated stands may promote specific microbiome assembly, potentially providing EM trees a competitive advantage.
Overall fungal community beta diversity may have been driven by mycorrhizal interactions with their environment. We did not find a decrease in fungal alpha diversity in EM-dominated stands relative to mixed AM-EM stands as has been reported in temperate forests (Eagar et al. 2021). Rather, fungal communities were more heterogeneous among EM-dominated stands than among mixed AM-EM stands. Greater fungal community heterogeneity across EM-dominated stands could have been driven by soil pH and fertility which varied across our four watersheds and can select for functionally distinct EM fungal communities (Corrales et al. 2016a). Specifically, low pH and fertility in Honda may select EM fungi that have a great capacity to alter C and N cycling through organic N uptake and that contribute abundant, low-quality fungal biomass to SOM pools (Seyfried et al. 2022). In contrast, relatively high soil pH and fertility in Alto Frio may select EM fungi which exclusively take up inorganic N and contribute limited, high-quality biomass to SOM pools (Seyfried et al. 2021a). These selective forces on EM fungi may represent a disproportionate influence on the overall fungal community based on the high relative abundance of EM fungi in these EM-dominated stands. In mixed AM-EM stands, environmental filtering for functionally robust, less specialized fungal communities may have driven greater homogeneity across sites despite differences in underlying soil pH and fertility (Kivlin et al. 2018). Shifts in fungal communities between stand mycorrhizal types were also correlated with higher relative abundance of EM fungi and lower relative abundance of fungal pathogens and saprotrophs in EM-dominated stands than in the AM-EM mixed stands. Interactions among fungal guilds may help promote positive plant-soil feedbacks associated with EM mutualisms (Bennett et al. 2017) by creating favorable microbiomes (i.e., lower potential pathogen loads and decomposer abundance) in EM-dominated forest stands. The functional composition of soil fungal communities in EM-dominated forest stands may partially drive the effects of EM trees on ecosystem function (McGuire et al. 2010) and be influenced by underlying soil pH and fertility.
The relationships between fungal and prokaryotic communities may highlight tradeoffs in belowground dynamics for different stand mycorrhizal types. Fungal and prokaryotic communities were more closely associated in EM-dominated than in mixed AM-EM stands. However, the divergence in this relationship (Procrustean residuals) increased with the relative abundance of fungal pathogens. Tradeoffs between defense, growth, nutrient acquisition, and mutualist collaboration in the development and morphology of root structures are an important component of plant development (Ravanbakhsh et al. 2019; Bergmann et al. 2020; Monson et al. 2022). For example, in tropical forests, the species most proficient at acquiring soil phosphorus are also the most vulnerable to pathogens (Laliberté et al. 2015; Lambers et al. 2018), with this tradeoff hypothesized to potentially help to maintain high plant diversity in these systems. Divergences between fungal and prokaryotic communities could be representative of tradeoffs made by plants to alter resource allocation for microbiome assembly in favor of pathogen protection. While this argument is largely speculative, the relationships we present here provide further support for microbial communities (or the capacity to manipulate them) as an extended plant root trait, which may be influenced by the surrounding environmental context (Freschet et al. 2021).
Conclusion
In a tropical montane forest, we demonstrate that EM-dominated stands are characterized by decreased prokaryotic diversity and different relative abundances of several important microbial functional groups compared to surrounding diverse mixed AM-EM forest. Differences in microbial communities between stand mycorrhizal types could be driven by overall differences in plant communities or microbiome assembly but likely contribute to broad EM-associated ecosystem effects. The relationships among different constituents of the soil microbiome could be an important extension of mycorrhizal function in response to the surrounding environment. Distinct soil microbiomes in EM-dominated versus mixed AM-EM stands may contribute to the effects of EM relationships on plant health or ecosystem function via changes to the relative abundance of fungal pathogens and saprotrophs. Overall, we found that the effects of mycorrhizal associations extend beyond the plant-fungal partnership into the broader soil microbiome and are especially pronounced for soil bacterial/archaeal communities.
References
Apprill A, McNally S, Parsons R, Weber L (2015) Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137. https://doi.org/10.3354/ame01753
Averill C, Hawkes CV (2016) Ectomycorrhizal fungi slow soil carbon cycling. Ecol Lett 19:937–947. https://doi.org/10.1111/ele.12631
Barceló M, van Bodegom PM, Tedersoo L, Olsson PA, Soudzilovskaia NA (2022) Mycorrhizal tree impacts on topsoil biogeochemical properties in tropical forests. J Ecol 110:1271–1282. https://doi.org/10.1111/1365-2745.13868
Barlow J, França F, Gardner TA, Hicks CC, Lennox GD, Berenguer E, Castello L, Economo EP, Ferreira J, Guénard B (2018) The future of hyperdiverse tropical ecosystems. Nature 559:517–526. https://doi.org/10.1038/s41586-018-0301-1
Bennett AE, Classen AT (2020) Climate change influences mycorrhizal fungal–plant interactions, but conclusions are limited by geographical study bias. Ecol 101:e02978. https://doi.org/10.1002/ecy.2978
Bennett JA, Maherali H, Reinhart KO, Lekberg Y, Hart MM, Klironomos J (2017) Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Sci 355:181–184. https://doi.org/10.1126/science.aai8212
Bergmann J, Weigelt A, van Der Plas TW, Laughlin DC, Kuyper TW, Guerrero-Ramirez N, Valverde-Barrantes OJ, Bruelheide H, Freschet GT, Iversen CM (2020) The fungal collaboration gradient dominates the root economics space in plants. Sci Adv 6:eaba3756. https://doi.org/10.1126/sciadv.aba3756
Berrios L, Yeam J, Holm L, Robinson W, Pellitier PT, Chin ML, Henkel TW, Peay KG (2023) Positive interactions between mycorrhizal fungi and bacteria are widespread and benefit plant growth. Curr Biol 33:2878–2887. https://doi.org/10.1016/j.cub.2023.06.010
Borowicz VA (2001) Do arbuscular mycorrhizal fungi alter plant–pathogen relations? Ecol 82:3057–3068. https://doi.org/10.2307/2679834
Callahan B, McMurdie P, Rosen M, Han A, Johnson A, Holmes S (2020) DADA2 ITS pipeline workflow (1.8). Retrieved September 18, 2020
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Chao A, Gotelli NJ, Hsieh T, Sander EL, Ma K, Colwell RK, Ellison AM (2014) Rarefaction and extrapolation with Hill numbers: a framework for sampling and estimation in species diversity studies. Ecol Monogr 84:45–67
Corrales A, Arnold AE, Ferrer A, Turner BL, Dalling JW (2016a) Variation in ectomycorrhizal fungal communities associated with Oreomunnea mexicana (Juglandaceae) in a Neotropical montane forest. Mycorrhiza 26:1–17. https://doi.org/10.1007/s00572-015-0641-8
Corrales A, Mangan SA, Turner BL, Dalling JW (2016b) An ectomycorrhizal nitrogen economy facilitates monodominance in a neotropical forest. Ecol Lett 19:383–392. https://doi.org/10.1111/ele.12570
Cronn R, Knaus BJ, Liston A, Maughan PJ, Parks M, Syring JV, Udall J (2012) Targeted enrichment strategies for next-generation plant biology. Am J Bot 99:291–311
Dalling JW, Turner BL (2021) Fortuna Forest Reserve, Panama: interacting effects of climate and soils on the biota of a wet premontane tropical forest. Smithsonian Scholarly Press
Delavaux CS, LaManna JA, Myers JA, Phillips RP, Aguilar S, Allen D, Alonso A, Anderson-Teixeira KJ, Baker ME, Baltzer JL (2023) Mycorrhizal feedbacks influence global forest structure and diversity. Commun Biol 6:1066
Dreischhoff S, Das IS, Jakobi M, Kasper K, Polle A (2020) Local responses and systemic induced resistance mediated by ectomycorrhizal fungi. Front Plant Sci 11:1908. https://doi.org/10.3389/fpls.2020.590063
Eagar AC, Mushinski RM, Horning AL, Smemo KA, Phillips RP, Blackwood CB (2021) Arbuscular mycorrhizal tree communities have greater soil fungal diversity and relative abundances of saprotrophs and pathogens than ectomycorrhizal tree communities. Appl Environ Microbiol 88:e01782-e11721. https://doi.org/10.1128/AEM.01782-21
Emmett BD, Lévesque-Tremblay V, Harrison MJ (2021) Conserved and reproducible bacterial communities associate with extraradical hyphae of arbuscular mycorrhizal fungi. ISME J 15:2276–2288. https://doi.org/10.1038/s41396-021-00920-2
Fernandez CW, Kennedy PG (2016) Revisiting the ʻGadgil effect’: do interguild fungal interactions control carbon cycling in forest soils? New Phytol 209:1382–1394. https://doi.org/10.1111/nph.13648
Fernandez CW, See CR, Kennedy PG (2019) Decelerated carbon cycling by ectomycorrhizal fungi is controlled by substrate quality and community composition. New Phytol 226(2):569–582. https://doi.org/10.1111/nph.16269
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364
Freschet GT, Roumet C, Comas LH, Weemstra M, Bengough AG, Rewald B, Bardgett RD, De Deyn GB, Johnson D, Klimešová J (2021) Root traits as drivers of plant and ecosystem functioning: current understanding, pitfalls and future research needs. New Phytol 232(2):1123–1158. https://doi.org/10.1111/nph.17072
Frey SD (2019) Mycorrhizal fungi as mediators of soil organic matter dynamics. Annu Rev Ecol Evol Syst 50:237–259. https://doi.org/10.1146/annurev-ecolsys-110617-062331
Hart TB, Hart JA, Murphy PG (1989) Monodominant and species-rich forests of the humid tropics: causes for their co-occurrence. Am Nat 133:613–633
Hawkes CV, DeAngelis KM, Firestone MK (2007) Root interactions with soil microbial communities and processes. Pages 1–29 The Rhizosphere. Elsevier
Heděnec P, Nilsson LO, Zheng H, Gundersen P, Schmidt IK, Rousk J, Vesterdal L (2020) Mycorrhizal association of common European tree species shapes biomass and metabolic activity of bacterial and fungal communities in soil. Soil Biol Biochem 149:107933
Heděnec P, Zheng H, Siqueira DP, Lin Q, Peng Y, Schmidt IK, Frøslev TG, Kjøller R, Rousk J, Vesterdal L (2023) Tree species traits and mycorrhizal association shape soil microbial communities via litter quality and species mediated soil properties. For Ecol Manage 527:120608. https://doi.org/10.1016/j.foreco.2022.120608
Hill MO (1973) Diversity and evenness: a unifying notation and its consequences. Ecol 54:427–432
Jacobs LM, Sulman BN, Brzostek ER, Feighery JJ, Phillips RP (2018) Interactions among decaying leaf litter, root litter and soil organic matter vary with mycorrhizal type. J Ecol 106:502–513. https://doi.org/10.1111/1365-2745.12921
Jose S, Gillespie AR (1998) Allelopathy in black walnut (Juglans nigra L.) alley cropping. II. Effects of juglone on hydroponically grown corn (Zea mays L.) and soybean (Glycine max L. Merr.) growth and physiology. Plant Soil 203:199–206
Jost L (2007) Partitioning diversity into independent alpha and beta components. Ecol 88:2427–2439
Kivlin SN, Bedoya R, Hawkes CV (2018) Heterogeneity in arbuscular mycorrhizal fungal communities may contribute to inconsistent plant-soil feedback in a Neotropical forest. Plant Soil 432:29–44
Laliberté E, Lambers H, Burgess TI, Wright SJ (2015) Phosphorus limitation, soil-borne pathogens and the coexistence of plant species in hyperdiverse forests and shrublands. New Phytol 206:507–521. https://doi.org/10.1111/nph.13203
Lambers H, Albornoz F, Kotula L, Laliberté E, Ranathunge K, Teste FP, Zemunik G (2018) How belowground interactions contribute to the coexistence of mycorrhizal and non-mycorrhizal species in severely phosphorus-impoverished hyperdiverse ecosystems. Plant Soil 424:11–33. https://doi.org/10.1007/s11104-017-3427-2
Li D (2018) hillR: taxonomic, functional, and phylogenetic diversity and similarity through Hill Numbers. J Open Source Softw 3:1041. https://doi.org/10.21105/joss.01041
Liang M, Johnson D, Burslem DF, Yu S, Fang M, Taylor JD, Taylor AF, Helgason T, Liu X (2020) Soil fungal networks maintain local dominance of ectomycorrhizal trees. Nat Commun 11:2636. https://doi.org/10.1038/s41467-020-16507-y
Liu Y, Sun Q, Li J, Lian B (2018) Bacterial diversity among the fruit bodies of ectomycorrhizal and saprophytic fungi and their corresponding hyphosphere soils. Sci Rep 8:11672. https://doi.org/10.1038/s41598-018-30120-6
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:1–21
MacArthur RH (1965) Patterns of species diversity. Biol Rev 40:510–533
McGuire KL, Fierer N, Bateman C, Treseder KK, Turner BL (2012) Fungal community composition in neotropical rain forests: the influence of tree diversity and precipitation. Microb Ecol 63:804–812
McGuire KL, Zak DR, Edwards IP, Blackwood CB, Upchurch R (2010) Slowed decomposition is biotically mediated in an ectomycorrhizal, tropical rain forest. Oecologia 164:785–795. https://doi.org/10.1038/s41598-018-30120-6
McMurdie PJ, Holmes S (2014) Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531
Monson RK, Trowbridge AM, Lindroth RL, Lerdau MT (2022) Coordinated resource allocation to plant growth–defense tradeoffs. New Phytol 233:1051–1066. https://doi.org/10.1111/nph.17773
Myrold DD (2021) Transformations of nitrogen. Pages 385–421 Principles and applications of soil microbiology. Elsevier
Nemergut DR, Cleveland CC, Wieder WR, Washenberger CL, Townsend AR (2010) Plot-scale manipulations of organic matter inputs to soils correlate with shifts in microbial community composition in a lowland tropical rain forest. Soil Biol Biochem 42:2153–2160
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG (2016) FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248. https://doi.org/10.1016/j.funeco.2015.06.006
Nilsson RH, Larsson K-H, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L (2019) The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res 47:D259–D264. https://doi.org/10.1093/nar/gky1022
Oh YM, Kim M, Lee-Cruz L, Lai-Hoe A, Go R, Ainuddin N, Rahim RA, Shukor N, Adams JM (2012) Distinctive bacterial communities in the rhizoplane of four tropical tree species. Microb Ecol 64:1018–1027. https://doi.org/10.1007/s00248-012-0082-2
Oksanen J, Blanchet FG, Kindt R, Legendre P, O’Hara R, Simpson GL, Solymos P, Stevens MHH, Wagner H (2010) Vegan: community ecology package. R package version 1.17–4
Parada AE, Needham DM, Fuhrman JA (2016) Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. https://doi.org/10.1111/1462-2920.13023
Peh KSH, Lewis SL, Lloyd J (2011) Mechanisms of monodominance in diverse tropical tree-dominated systems. J Ecol 99:891–898. https://doi.org/10.1111/j.1365-2745.2011.01827.x
Pellegrin C, Morin E, Martin FM, Veneault-Fourrey C (2015) Comparative analysis of secretomes from ectomycorrhizal fungi with an emphasis on small-secreted proteins. Front Microbiol 6:1278
Pellitier PT, Zak DR (2018) Ectomycorrhizal fungi and the enzymatic liberation of nitrogen from soil organic matter: why evolutionary history matters. New Phytol 217:68–73. https://doi.org/10.1111/nph.14598
Phillips RP, Brzostek E, Midgley MG (2013) The mycorrhizal-associated nutrient economy: a new framework for predicting carbon-nutrient couplings in temperate forests. New Phytol 199:41–51. https://doi.org/10.1111/nph.12221
Prada CM, Morris A, Andersen KM, Turner BL, Caballero P, Dalling JW (2017) Soils and rainfall drive landscape-scale changes in the diversity and functional composition of tree communities in premontane tropical forest. J Veg Sci 28:859–870. https://doi.org/10.1111/jvs.12540
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596
R Core Team R (2013) R: a language and environment for statistical computing
Rambelli A (1973) The rhizosphere of mycorrhizae. Ectomycorrhizae 299–343
Ravanbakhsh M, Kowalchuk GA, Jousset A (2019) Root-associated microorganisms reprogram plant life history along the growth–stress resistance tradeoff. ISME J 13:3093–3101. https://doi.org/10.1038/s41396-019-0501-1
Revillini D, David AS, Reyes AL, Knecht LD, Vigo C, Allen P, Searcy CA, Afkhami ME (2023) Allelopathy-selected microbiomes mitigate chemical inhibition of plant performance. New Phytol 240(5):2007–2019. https://doi.org/10.1111/nph.19249
Revillini D, Gehring CA, Johnson NC (2016) The role of locally adapted mycorrhizas and rhizobacteria in plant–soil feedback systems. Funct Ecol 30:1086–1098
Sangwan S, Prasanna R (2022) Mycorrhizae helper bacteria: unlocking their potential as bioenhancers of plant–arbuscular mycorrhizal fungal associations. Microb Ecol 84:1–10
Seyfried GS, Canham CD, Dalling JW, Yang WH (2021a) The effects of tree-mycorrhizal type on soil organic matter properties from neighborhood to watershed scales. Soil Biol Biochem 161:108385. https://doi.org/10.1016/j.soilbio.2021.108385
Seyfried GS, Corrales A, Kent AD, Dalling JW, Yang WH (2022) Watershed-scale variation in potential fungal community contributions to ectomycorrhizal biogeochemical syndromes. Ecosyst 26(4):724–739. https://doi.org/10.1007/s10021-022-00788-z
Seyfried GS, Dalling JW, Yang WH (2021b) Mycorrhizal type effects on leaf litter decomposition depend on litter quality and environmental context. Biogeochem 155(1):21–38. https://doi.org/10.1007/s10533-021-00810-x
Smith SE, Smith FA (2011) Roles of arbuscular mycorrhizas in plant nutrition and growth: new paradigms from cellular to ecosystem scales. Annu Rev Plant Biol 62:227–250. https://doi.org/10.1146/annurev-arplant-042110-103846
Suriyavirun N, Krichels AH, Kent AD, Yang WH (2019) Microtopographic differences in soil properties and microbial community composition at the field scale. Soil Biol Biochem 131:71–80
Tarkka MT, Drigo B, Deveau A (2018) Mycorrhizal microbiomes. Mycorrhiza 28:403–409
Torti SD, Coley PD, Kursar TA (2001) Causes and consequences of monodominance in tropical lowland forests. Am Nat 157:141–153. https://doi.org/10.1086/318629
Turner BL, Dalling JW (2021) Soils of the Fortuna forest reserve. In: Dalling JW, Turner BL (eds) Fortuna Forest Reserve, Panama: interacting effects of climate and soils on the biota of a wet premontane tropical forest. Smithsonian Scholarly Press, Washington, D.C, pp 47–137
Uroz S, Courty PE, Oger P (2019) Plant symbionts are engineers of the plant-associated microbiome. Trends Plant Sci 24:905–916
van Der Heijden MG, Martin FM, Selosse MA, Sanders IR (2015) Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol 205:1406–1423. https://doi.org/10.1111/nph.13288
Veresoglou SD, Rillig MC (2012) Suppression of fungal and nematode plant pathogens through arbuscular mycorrhizal fungi. Biol Let 8:214–217
Wang L, Zhang L, George TS, Feng G (2023) A core microbiome in the hyphosphere of arbuscular mycorrhizal fungi has functional significance in organic phosphorus mineralization. New Phytol 238:859–873. https://doi.org/10.1111/nph.18642
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: a Guide to Methods and Applications 18:315–322
Xu Y, Chen Z, Li X, Tan J, Liu F, Wu J (2023) Mycorrhizal fungi alter root exudation to cultivate a beneficial microbiome for plant growth. Funct Ecol 37:664–675. https://doi.org/10.1111/1365-2435.14249
Zak DR, Holmes WE, White DC, Peacock AD, Tilman D (2003) Plant diversity, soil microbial communities, and ecosystem function: are there any links? Ecol 84:2042–2050
Zhang L, Zhou J, George TS, Limpens E, Feng G (2022) Arbuscular mycorrhizal fungi conducting the hyphosphere bacterial orchestra. Trends Plant Sci 27(2):402–411. https://doi.org/10.1016/j.tplants.2021.10.008
Acknowledgements
We would like to thank Kristin Saltonstall for the use of the lab space, as well as Evidelio Garcia and Carlos Espinosa for field assistance. The Smithsonian Tropical Research Institute provided logistical support at the Fortuna Forest Reserve. AHK is supported by the USDA Forest Service Rocky Mountain Research Station; the findings and conclusions in this publication are those of the authors and should not be construed to represent any official USDA or US Government determination or policy.
Funding
Joseph D. Edwards was supported by the Cooperative State Research, Education, and Extension Service, US Department of Agriculture, under project number ILLU 875–952, as well as by US National Science Foundation Division of Environmental Biology award #2305863. Alexander H. Krichels and Georgia S. Seyfried were supported by The National Science Foundation Integrative Graduate Education and Research Traineeship Program (NSF IGERT #1069157).
Author information
Authors and Affiliations
Contributions
James Dalling maintains research sites; Alex H. Krichels and Wendy H. Yang collected samples; Alex H. Krichels and Angela D. Kent extracted and sequenced DNA; Joseph D. Edwards analyzed data, generated figures, and wrote the main text of the manuscript. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Edwards, J.D., Krichels, A.H., Seyfried, G.S. et al. Soil microbial community response to ectomycorrhizal dominance in diverse neotropical montane forests. Mycorrhiza 34, 95–105 (2024). https://doi.org/10.1007/s00572-023-01134-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-023-01134-4