Abstract
Key message
This study developed and validated a set of polymorphic and cross-transferable EST-SSR markers in pecan. 31 polymorphic primers were qualified for genetic diversity evaluation.
Abstract
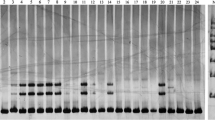
Pecan is a famous nut tree with high nutritional value. Nowadays, few EST-SSR markers have been developed for this species. Here, 31,074 protein-coding genes of pecan with 46.43 Mb were employed for EST-SSR identification. In total, 9522 EST-SSR loci were detected from 5752 genes with a distribution frequency of 16.69% of total genes and an average density of one SSR per 4.88 kb. Among them, the di-nucleotide (44.24%) was the most abundant motif, followed by mono-, tri-, tetra-, hexa-, and penta-nucleotide repeats. In addition, a total of 7764 EST-SSR loci were distributed in UTR regions, while the rest of the 1758 were located at CDS regions. Based on the flanked sequences surrounding SSR loci, 3586 primers were successfully developed and 250 pairs were randomly selected for verification using six pecan cultivars. After amplification, 171 (68.40%) primer pairs yielded the expected bands, of which, 159 showed polymorphisms. Among the polymorphic primers, 76 pairs could be cross-amplified in four other Carya species (Carya. laciniosa, C. ovata, C. hunanensis, and C. cathayensis). To further characterize the usefulness of EST-SSRs, 31 polymorphic primers were subjected to assess the genetic diversity and relationship of 40 pecan lines, in which the mean number of alleles per locus and polymorphism information content were 6.90 and 0.70, respectively, suggesting the capacity of the markers to monitor high level of informativeness. Cluster analysis indicated that some accessions in the corresponding subgroup were in good agreement with their genetic backgrounds and morphological traits. The EST-SSR resources offered here constitute a valuable tool for genetic diversity analysis in pecan.



Similar content being viewed by others
References
Alonso-Blanco C, Mendez-Vigo B, Koornneef M (2004) From phenotypic to molecular polymorphisms involved in naturally occurring variation of plant development. Int J Dev Biol 49:717–732
Babayeva S, Akparov Z, Abbasov M, Mammadov A, Zaifizadeh M, Street K (2009) Diversity analysis of Central Asia and Caucasian lentil (Lens culinaris Medik.) germplasm using SSR fingerprinting. Genet Resour Crop Ev 56:293–298
Bazzo BR, de Carvalho LM, Carazzolle MF, Pereira GAG, Colombo CA (2018) Development of novel EST-SSR markers in the macaúba palm (Acrocomia aculeata) using transcriptome sequencing and cross-species transferability in Arecaceae species. BMC Plant Biol 18:1–10
Beier S, Thiel T, Münch T, Scholz U, Mascher M (2017) MISA-web: a web server for microsatellite prediction. Bioinformatics 33:2583–2585
Bentley N, Grauke L, Klein P (2019) Genotyping by sequencing (GBS) and SNP marker analysis of diverse accessions of pecan (Carya illinoinensis). Tree Genet Genomes 15:1–17
Bentley N, Grauke L, Ruhlman E, Klein RR, Kubenka K, Wang X, Klein P (2020) Linkage mapping and QTL analysis of pecan (Carya illinoinensis) full-siblings using genotyping-by-sequencing. Tree Genet Genomes 16:1–20
Bisen A, Khare D, Nair P, Tripathi N (2015) SSR analysis of 38 genotypes of soybean (Glycine Max (L.) Merr.) genetic diversity in India. Physiol Mol Biol Pla 21:109–115
Casales FG, Van der Watt E, Coetzer GM (2018) Propagation of pecan (Carya illinoensis): a review. Afr J Biotechnol 17:586–605
Cerna-Cortés JF, Simpson J, Martínez O, Martínez-Peniche RA (2003) Measurement of genetic diversity of native pecan [Carya illinoensis (Wangenh) K. Koch] populations established in Central Mexico and correlation with dichogamous flowering using AFLP. J Food Agric Environ 1:168–173
Cheng Y, Yang Y, Wang Z, Qi B, Yin Y, Li H (2015) Development and characterization of EST-SSR markers in Taxodium ‘zhongshansa.’ Plant Mol Biol Rep 33:1804–1814
Conner PJ, Wood BW (2001) Identification of pecan cultivars and their genetic relatedness as determined by randomly amplified polymorphic DNA analysis. J Am Soc Hortic Sci 126:474–480
Dang M, Zhang T, Hu Y, Zhou H, Woeste KE, Zhao P (2016) De novo assembly and characterization of bud, leaf and flowers transcriptome from Juglans regia L. for the identification and characterization of new EST-SSRs. Forests 7:247
Dillon NL, Innes DJ, Bally IS, Wright CL, Devitt LC, Dietzgen RG (2014) Expressed sequence tag-simple sequence repeat (EST-SSR) marker resources for diversity analysis of mango (Mangifera indica L.). Diversity 6:72–87
Dong M, Wang Z, He Q, Zhao J, Fan Z, Zhang J (2018) Development of EST-SSR markers in Larix principis-rupprechtii Mayr and evaluation of their polymorphism and cross-species amplification. Trees-Struct Funct 32:1559–1571
Durand J, Bodénès C, Chancerel E, Frigerio JM, Vendramin G, Sebastiani F, Buonamici A, Gailing O, Koelewijn HP, Villani F (2010) A fast and cost-effective approach to develop and map EST-SSR markers: oak as a case study. BMC Genomics 11:1–13
Feng S, Li W, Huang H, Wang J, Wu Y (2009) Development, characterization and cross-species/genera transferability of EST-SSR markers for rubber tree (Hevea brasiliensis). Mol Breeding 23:85–97
Garrido-Cardenas JA, Mesa-Valle C, Manzano-Agugliaro F (2018) Trends in plant research using molecular markers. Planta 247:543–557
Grauke L, Mendoza-Herrera M (2012) Population structure in the genus Carya. Acta Hortic. https://doi.org/10.17660/ActaHortic.2012.948.16
Grauke L, Iqbal MJ, Reddy AS, Thompson TE (2003) Developing microsatellite DNA markers in pecan. J Am Soc Hortic Sci 128:374–380
He X, Zheng J, Tian X, Jiao Z, Dou Q (2021) Genetic relationship analysis and fingerprint construction of Carya illinoensis varieties. Forest Res 34:95–102
Hirt H (1997) Multiple roles of MAP kinases in plant signal transduction. Trends Plant Sci 2:11–15
Hu J, Wang L, Li J (2011) Comparison of genomic SSR and EST-SSR markers for estimating genetic diversity in cucumber. Biol Plantarum 55:577–580
Hu XG, Liu H, Jin YQ, Sun YQ, Li Y, Zhao W, El-Kassaby YA, Wang XR, Mao JF (2016) De novo transcriptome assembly and characterization for the widespread and stress-tolerant conifer Platycladus orientalis. PLoS ONE 11:e0148985
Huang Y, Xiao L, Zhang Z, Zhang R, Wang Z, Huang C, Huang R, Luan Y, Fan T, Wang J (2019) The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition. GigaScience 8:giz036
Jia XD, Wang T, Zhai M, Li YR, Guo ZR (2011) Genetic diversity and identification of Chinese-grown pecan using ISSR and SSR markers. Molecules 16:10078–10092
Jia X, Deng Y, Sun X, Liang L, Su J (2016) De novo assembly of the transcriptome of Neottopteris nidus using Illumina paired-end sequencing and development of EST-SSR markers. Mol Breeding 36:94
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan A (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177:309–334
Kumpatla SP, Mukhopadhyay S (2005) Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species. Genome 48:985–998
Lacape JM, Dessauw D, Rajab M, Noyer JL, Hau B (2007) Microsatellite diversity in tetraploid Gossypium germplasm: assembling a highly informative genotyping set of cotton SSRs. Mol Breeding 19:45–58
Lee HY, Ro NY, Jeong HJ, Kwon JK, Jo J, Ha Y, Jung A, Han JW, Venkatesh J, Kang BC (2016) Genetic diversity and population structure analysis to construct a core collection from a large Capsicum germplasm. BMC Genet 17:142
Li Y, Jia LK, Zhang FQ, Wang ZH, Chen SL, Gao QB (2019) Development of EST-SSR markers in Saxifraga sinomontana (Saxifragaceae) and cross-amplification in three related species. Appl Plant Sci 7:e11269
Li X, Liu X, Wei J, Li Y, Tigabu M, Zhao X (2020) Development and transferability of EST-SSR markers for Pinus koraiensis from cold-stressed transcriptome through Illumina Sequencing. Genes 11:500
Liu SR, Li WY, Long D, Hu CG, Zhang JZ (2013) Development and characterization of genomic and expressed SSRs in citrus by genome-wide analysis. PLoS ONE 8:e75149
Lovell JT, Bentley NB, Bhattarai G, Jenkins JW, Sreedasyam A, Alarcon Y, Bock C, Boston LB, Carlson J, Cervantes K (2021) Four chromosome scale genomes and a pan-genome annotation to accelerate pecan tree breeding. Nat Commun 12:4125
Luo L, Yang Y, Zhao H, Leng P, Hu Z, Wu J, Zhang K (2021) Development of EST-SSR markers and association analysis of floral scent in tree peony. Sci Hortic-Amsterdam 289:110409
Matsuda F, Nakabayashi R, Yang Z, Okazaki Y, Ji Y, Ebana K, Yano M, Saito K (2015) Metabolome-genome-wide association study dissects genetic architecture for generating natural variation in rice secondary metabolism. Plant J 81:13–23
Metzgar D, Bytof J, Wills C (2000) Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res 10:72–80
Mo Z, Chen Y, Lou W, Jia X, Zhai M, Xuan J, Guo Z, Li Y (2020a) Identification of suitable reference genes for normalization of real-time quantitative PCR data in pecan (Carya illinoinensis). Trees-Struct Funct 34:1233–1241
Mo Z, Lou W, Chen Y, Jia X, Zhai M, Guo Z, Xuan J (2020b) The chloroplast genome of Carya illinoinensis: genome structure, adaptive evolution, and phylogenetic analysis. Forests 11:207
Moccia MD, Oger-Desfeux C, Marais GA, Widmer A (2009) A White Campion (Silene latifolia) floral expressed sequence tag (EST) library: annotation, EST-SSR characterization, transferability, and utility for comparative mapping. BMC Genomics 10:243
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30:194–200
Nandha PS, Singh J (2014) Comparative assessment of genetic diversity between wild and cultivated barley using g SSR and EST-SSR markers. Plant Breeding 133:28–35
Nyabera LA, Nzuki IW, Runo SM, Amwayi PW (2021) Assessment of genetic diversity of pumpkins (Cucurbita spp.) from western Kenya using SSR molecular markers. Mol Biol Rep 48:2253–2260
Palmieri DA, Novelli VM, Bastianel M, Cristofani-Yaly M, Astúa-Monge G, Carlos EF, Oliveira ACd, Machado MA (2007) Frequency and distribution of microsatellites from ESTs of citrus. Genet Mol Biol 30:1009–1018
Pan L, Huang T, Yang Z, Tang L, Cheng Y, Wang J, Ma X, Zhang X (2018) EST-SSR marker characterization based on RNA-sequencing of Lolium multiflorum and cross transferability to related species. Mol Breeding 38:80
Pandey MK, Gautami B, Jayakumar T, Sriswathi M, Upadhyaya HD, Gowda MVC, Radhakrishnan T, Bertioli DJ, Knapp SJ, Cook DR (2012) Highly informative genic and genomic SSR markers to facilitate molecular breeding in cultivated groundnut (Arachis hypogaea). Plant Breeding 131:139–147
Parthiban S, Govindaraj P, Senthilkumar S (2018) Comparison of relative efficiency of genomic SSR and EST-SSR markers in estimating genetic diversity in sugarcane. 3 Biotech 8:144
Poczai P, Varga I, Laos M, Cseh A, Bell N, Valkonen J, Hyvönen J (2013) Advances in plant gene-targeted and functional markers: a review. Plant Methods 9:6
Poletto T, Poletto I, Silva LMM, Muniz MFB, Reiniger LRS, Richards N, Stefenon VM (2020) Morphological, chemical and genetic analysis of southern Brazilian pecan (Carya illinoinensis) accessions. Sci Hortic-Amsterdam 261:108863
Qi F, Zhang F (2020) Cell cycle regulation in the plant response to stress. Front Plant Sci 10:1765
Qiu L, Yang C, Tian B, Yang JB, Liu A (2010) Exploiting EST databases for the development and characterization of EST-SSR markers in castor bean (Ricinus communis L.). BMC Plant Biol 10:278
Şelale H, Çelik I, Gültekin V, Allmer J, Doğanlar S, Frary A (2013) Development of EST-SSR markers for diversity and breeding studies in opium poppy. Plant Breeding 132:344–351
Sun H, Liu Y, Gai Y, Geng J, Chen L, Liu H, Kang L, Tian Y, Li Y (2015) De novo sequencing and analysis of the cranberry fruit transcriptome to identify putative genes involved in flavonoid biosynthesis, transport and regulation. BMC Genomics 16:652
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Wang L, Gong X, Jin L, Li H, Lu J, Duan J, Ma L (2019) Development and validation of EST-SSR markers of Magnolia wufengensis using de novo transcriptome sequencing. Trees-Struct Funct 33:1213–1223
Wang L, Zhang R, Geng M, Qin Y, Liu H, Li L, Li M (2021) De novo transcriptome assembly and EST-SSR markers development for Zelkova schneideriana Hand. –Mazz. (Ulmaceae). Biotech 11:420
Wang X, Chatwin W, Hilton A, Kubenka K (2022) Genetic diversity revealed by microsatellites in genus Carya. Forests 13:188
Wood BW, Payne JA, Grauke LJ (1990) The rise of the US pecan industry. HortScience 25:594–723
Wu J, Cai C, Cheng F, Cui H, Zhou H (2014) Characterisation and development of EST-SSR markers in tree peony using transcriptome sequences. Mol Breeding 34:1853–1866
Xia W, Xiao Y, Liu Z, Luo Y, Mason AS, Fan H, Yang Y, Zhao S, Peng M (2014) Development of gene-based simple sequence repeat markers for association analysis in Cocos nucifera. Mol Breeding 34:525–535
Xiao L, Yu M, Zhang Y, Hu J, Zhang R, Wang J, Guo H, Zhang H, Guo X, Deng T (2021) Chromosome-scale assembly reveals asymmetric paleo-subgenome evolution and targets for the acceleration of fungal resistance breeding in the nut crop, pecan. Plant Commun 2:100247
Xu X, Zhou C, Zhang Y, Zhang W, Gan X, Zhang H, Guo Y, Gan S (2018) A novel set of 223 EST-SSR markers in Casuarina L. ex Adans.: polymorphisms, cross-species transferability, and utility for commercial clone genotyping. Tree Genet Genomes 14:30
Yang Y, He R, Zheng J, Hu Z, Wu J, Leng P (2020) Development of EST-SSR markers and association mapping with floral traits in Syringa oblata. BMC Plant Biol 20:436
Yoruk B, Taskin V (2014) Genetic diversity and relationships of wild and cultivated olives in Turkey. Plant Syst Evol 300:1247–1258
Zhai M, Jia XD, Zhang JY, Guo ZR, Xuan JP (2021) Characterization of the complete chloroplast genome of Carya laciniosa (F. Michx.) G. Don (Juglandaceae). Mitochondrial DNA B 6:1261–1262
Zhang MY, Fan L, Liu QZ, Song Y, Wei SW, Zhang SL, Wu J (2014) A novel set of EST-derived SSR markers for pear and cross-species transferability in Rosaceae. Plant Mol Biol Rep 32:290–302
Zhang Y, Zhang M, Hu Y, Zhuang X, Xu W, Li P, Wang Z (2019) Mining and characterization of novel EST-SSR markers of Parrotia subaequalis (Hamamelidaceae) from the first Illumina-based transcriptome datasets. PLoS ONE 14:e0215874
Zhang C, Yao X, Ren H, Chang J, Wu J, Shao W, Fang Q (2020) Characterization and development of genomic SSRs in Pecan (Carya illinoinensis). Forests 11:61
Zhao Y, Williams R, Prakash C, He G (2012) Identification and characterization of gene-based SSR markers in date palm (Phoenix dactylifera L.). BMC Plant Biol 12:237
Zhou Q, Luo D, Ma L, Xie W, Wang Y, Wang Y, Liu Z (2016) Development and cross-species transferability of EST-SSR markers in Siberian wildrye (Elymus sibiricus L.) using Illumina sequencing. Sci Rep 6:20549
Zhou Q, Zhou PY, Zou WT, Li YG (2021) EST-SSR marker development based on transcriptome sequencing and genetic analyses of Phoebe bournei (Lauraceae). Mol Biol Rep 48:2201–2208
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 31901347), the Innovation Capacity Construction Project of Jiangsu Province (BM2018022), and Jiangsu Provinces Innovation and Introduction Project of Forestry Science and Technology (LYKJ[2020]04).
Author information
Authors and Affiliations
Contributions
ZM and JX: conceived and designed the research. WL: performed the experiments and drafted the manuscript; YL, LH, and XY: collected the sampling materials; MZ: analyzed the data; all authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Luis Gómez.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lou, W., Lyu, Y., Hu, L. et al. Development and characterization of EST-SSR markers in pecan (Carya illinoinensis). Trees 37, 297–307 (2023). https://doi.org/10.1007/s00468-022-02347-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-022-02347-4