Abstract
Key message
This work contributes to the identification and understanding of CBF genes and their putative role in the mechanisms of cold tolerance in eucalypt species.
Abstract
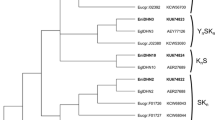
Three new CBF genes were isolated from E. globulus-denominated EglCBF1a, c, and d, coding for proteins of 220, 229, and 196 amino acids, respectively. The sequence analysis showed that the three predicted proteins contain an AP2 DNA-binding domain and two CBF signature sequences. Phylogenetic analysis demonstrated that these proteins were highly similar to those described in E. grandis and E. gunnii. Transcript abundance analysis in three different E. globulus genotypes exposed to a cold acclimation treatment showed that these CBF genes were highly related to the acclimation process and presented the highest relative expression at freezing temperatures. EglCBF1a showed the highest expression level (1311-fold change) in the cold-tolerant genotype (R1). EglCBF1a and d genes were induced by chilling and freezing temperatures, while EglCBF1c was constitutively expressed, increasing its transcript level when plants were exposed to freezing temperatures. The constitutive overexpression of each E. globulus CBF gene in Arabidopsis plants induces the endogenous CBF regulon gene expression of Arabidopsis and enhanced its tolerance to freezing, with additional phenotypic effects including growth inhibition and delayed flowering. These results indicate that the three EglCBF genes analyzed play important roles under cold acclimation processes in E. globulus and are involved in the signaling pathway of cold stress and the freezing tolerance phenotype observed on specific genotypes of this species and in transgenic Arabidopsis lines.







Similar content being viewed by others
References
Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P (2008) The cold-inducible CBF1 factor–dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20(8):2117–2129
Almeida M, Chaves M, Silva J (1994) Cold acclimation in eucalypt hybrids. Tree Physiol 14(7-8-9):921–932
Azar S, SanClemente H, Marque G, Dunand C, Marque C, Teulières C (2011) Bioinformatic prediction of the AP2/ERF family genes in Eucalyptus grandis: focus on the CBF family. BMC Proc 5 (Suppl 7):P165
Benedict C, Jeffrey S, Rengong M, Yongjian C, Rishikesh B, Norman P, Chad E, Tony H, Vaughan H (2006) The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Env 29(7):1259–1272
Bonawitz N, Soltau W, Blatchley M, Powers B, Hurlock A, Seals L, Weng J-K, Stout J, Chapple C (2012) REF4 and RFR1, subunits of the transcriptional coregulatory complex mediator, are required for phenylpropanoid homeostasis in Arabidopsis. J Biol Chem 287(8):5434–5445
Byun M, Lee J, Cui L, Kang Y, Oh T, Park H, Lee H, Kim W (2015) Constitutive expression of DaCBF7, an Antarctic vascular plant Deschampsia antarctica CBF homolog, resulted in improved cold tolerance in transgenic rice plants. Plant Sci 236:61–74
Canella D, Gilmour S, Kuhn L, Thomashow M (2010) DNA binding by the Arabidopsis CBF1 transcription factor requires the PKKP/RAGRxKFxETRHP signature sequence. Biochim et Biophy Acta (BBA) Gene Regul Mech 1799(5–6):454–462
Cao PB, Azar S, SanClemente H, Mounet F, Dunand C, Marque G, Marque C, Teulières C (2015) Genome-wide analysis of the AP2/ERF family in eucalyptus grandis: an intriguing over-representation of stress-responsive DREB1/CBF genes. PLoS One 10(4):e0121041
Champ K, Febres V, Moore G (2007) The role of CBF transcriptional activators in two Citrus species (Poncirus and Citrus) with contrasting levels of freezing tolerance. Physiol Plant 129(3):529–541
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11(2):113–116
Chaves M, Maroco J, Pereira J (2003) Understanding plant responses to drought—from genes to the whole plant. Funct Plant Biol 30(3):239–264
Chinnusamy V, Zhu J-K, Sunkar R (2010) Gene regulation during cold stress acclimation in plants. Methods Mol Biol 639:39–55
Clough S, Bent A (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743
Curtis M, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133(2):462–469
El Kayal W, Navarro M, Marque G, Keller G, Marque C, Teulières C (2006) Expression profile of CBF-like transcriptional factor genes from Eucalyptus in response to cold. J Exp Bot 57(10):2455–2469
Fang Z, Zhang X, Gao J, Wang P, Xu X, Liu Z, Shen S, Feng B (2015) A Buckwheat (Fagopyrum esculentum) DRE-Binding transcription factor gene, FeDREB1, enhances freezing and drought tolerance of transgenic Arabidopsis. Plant Mol Biol Rep 33(5):1510–1525
FAO (2007) State of the World’s Forests 2007. Electronic Publishing Policy and Support Branch, Communication Division, Rome
Fernández M, Villarroel C, Balbontín C, Valenzuela S (2010) Validation of reference genes for real-time qRT-PCR normalization during cold acclimation in Eucalyptus globulus. Trees Struct Funct 24 (6):1109–1116
Fernández M, Valenzuela S, Arora R, Chen K (2012) Isolation and characterization of three cold acclimation-responsive dehydrin genes from Eucalyptus globulus. Tree Genet Genom 8(1):149–162
Fernández M, Troncoso V, Valenzuela S (2015) Transcriptome profile in response to frost tolerance in Eucalyptus globulus. Plant Mol Biol Rep 33(5):1472–1485
Gamboa M, Rasmussen-Poblete S, Valenzuela P, Krauskopf E (2007) Isolation and characterization of a cDNA encoding a CBF transcription factor from E. globulus. Plant Physiol Biochem 45(1):1–5
Geldres E, Schlatter J (2004) Crecimiento de las plantaciones de Eucalyptus globulus sobre suelos rojo arcillosos de la provincia de Osorno, Décima Región. Bosque (Valdivia) 25:95–101
Gilmour S, Zarka D, Stockinger E, Salazar M, Houghton J, Thomashow M (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16(4):433–442
Gilmour S, Fowler S, Thomashow M (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol 54(5):767–781
Grattapaglia D (2004) Integrating genomics into Eucalyptus breeding. Genet Mol Res 3(3):369–379
Haake V, Cook D, Riechmann J, Pineda O, Thomashow M, Zhang J (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130(2):639–648
Harrison S, Mott E, Parsley K, Aspinall S, Gray J, Cottage A (2006) A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant. Methods 2(1):19
Iglesias-Trabado G, Wilstermann D (2009) Eucalyptus universalis. Global cultivated eucalypt forests map 2009. Version 1.0.2 In GIT Forestry Consulting’s EUCALYPTOLOGICS: Information resources on Eucalyptus cultivation worldwide. XIII World Forestry Congress, Argentina Mar 29th 2009. Retrieved from http://www.git-forestry.com
INFOR (2014) Anuario forestal. Instituto forestal: Boletín estadísico Nº144 (1):159p http://wef.infor.cl/publicaciones/publicaciones.php. Accessed 27 July 2016
Jaglo K, Kleff S, Amundsen K, Zhang X, Haake V, Zhang J, Deits T, Thomashow M (2001) Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol 127(3):910–917
Jaglo-Ottosen K, Gilmour S, Zarka D, Schabenberger O, Thomashow M (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280(5360):104–106
Jofuku K, den Boer B, Van Montagu M, Okamuro J (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6(9):1211–1225
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotech 17(3):287–291
Kim S, Nam K (2010) Physiological roles of ERD10 in abiotic stresses and seed germination of Arabidopsis. Plant Cell Rep 29(2):203–209
Kim Y, Lee M, Lee J-H, Lee H-J, Park C-M (2015) The unified ICE–CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol Biol 89(1):187–201
Levitt J (1980) Responses of Plants to Environmental Stresses, vol 1. Academic Press, New York, NY
Li J, Wang N, Xin H, Li S (2013) Overexpression of VaCBF4, a transcription factor from Vitis amurensis, improves cold tolerance accompanying increased resistance to drought and salinity in Arabidopsis. Plant Mol Biol Rep 31(6):1518–1528
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell Online 10(8):1391–1406
Livak K, Schmittgen T (2001) Analysis of relative gene expression data using real-time quantitative pcr and the 2– ∆∆Ct method. Methods 25(4):402–408
Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2004) Dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J 37(5):720–729
Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2008) The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J 56(4):613–626
Mäntylä E, Lång V, Palva ET (1995) Role of abscisic acid in drought-induced freezing tolerance, cold acclimation, and accumulation of LT178 and RAB18 proteins in Arabidopsis thaliana. Plant Physiol 107(1):141–148
McGinnis K, Thomas S, Soule J, Strader L, Zale J, Sun T, Steber C (2003) The Arabidopsis sleepy1 gene encodes a putative f-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15(5):1120–1130
Medina J, Catalá R, Salinas J (2011) The CBFs: Three Arabidopsis transcription factors to cold acclimate. Plant Sci 180(1):3–11
Navarro M, Marque G, Ayax C, Keller G, Borges J, Marque C, Teulières C (2009) Complementary regulation of four Eucalyptus CBF genes under various cold conditions. J Exp Bot 60(9):2713–2724
Navarro M, Ayax C, Martinez Y, Laur J, El Kayal W, Marque C, Teulières C (2011) Two EguCBF1 genes overexpressed in Eucalyptus display a different impact on stress tolerance and plant development. Plant Biotechnol J 9(1):50–63
Novillo F, Alonso JM, Ecker JR, Salinas J (2004) CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 101(11):3985–3990
Novillo F, Medina J, Salinas J (2007) Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc Natl Acad Sci 104(52):21002–21007
Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7(2):173–182
Peng Y-L, Wang Y-S, Cheng H, Sun C-C, Wu P, Wang L-Y, Fei J (2013) Characterization and expression analysis of three CBF/DREB1 transcriptional factor genes from mangrove Avicennia marina. Aquat Toxicol 140–141:68–76
Pita P, Pardos JA (2001) Growth, leaf morphology, water use and tissue water relations of Eucalyptus globulus clones in response to water deficit. Tree Physiol 21(9):599–607
Riechmann J, Meyerowitz E (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379:633–646
Rodziewicz P, Swarcewicz B, Chmielewska K, Wojakowska A, Stobiecki M (2014) Influence of abiotic stresses on plant proteome and metabolome changes. Acta Physiologiae Plantarum 36(1):1–19
Ruelland E, Vaultier M-N, Zachowski A, Hurry V (2009) Chap. 2 Cold signalling and cold acclimation in plants. In: Advances in botanical research, Vol 49. Academic Press, Cambridge, pp 35–150
Ryu J, Hong S-Y, Jo S-H, Woo J-C, Lee S, Park C-M (2014) Molecular and functional characterization of cold-responsive C-repeat binding factors from Brachypodium distachyon. BMC Plant Biol 14(1):15
Sakuma Y, Liu Q, Dubouzet J, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-Binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290(3):998–1009
Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13(1):61–72
Siddiqua M, Nassuth A (2011) Vitis CBF1 and Vitis CBF4 differ in their effect on Arabidopsis abiotic stress tolerance, development and gene expression. Plant Cell Env 34(8):1345–1359
Stockinger E, Gilmour S, Thomashow M (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci 94(3):1035–1040
Tang M, Lü S, Jing Y, Zhou X, Sun J, Shen S (2005) Isolation and identification of a cold-inducible gene encoding a putative DRE-binding transcription factor from Festuca arundinacea. Plant Physiol Biochem 43(3):233–239
Thomashow M (1990) Molecular Genetics of Cold Acclimation in Higher Plants. In: John GS (ed) Advances in genetics, Vol 28. Academic Press, Cambridge, pp 99–131
Thomashow M (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50(1):571–599
Thomashow M (2010) Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiol 154(2):571–577
Thomashow M, Gilmour S, Stockinger E, Jaglo-Ottosen K, Zarka D (2001) Role of the Arabidopsis CBF transcriptional activators in cold acclimation. Physiol Plant 112(2):171–175
Tibbits W, White T, Hodge G, Borralho N (2006) Genetic variation in frost resistance of Eucalyptus globulus ssp. globulus assessed by artificial freezing in winter. Aust J Bot 54(6):521–529
Tillett R, Wheatley M, Tattersall E, Schlauch K, Cramer G, Cushman J (2012) The Vitis vinifera C-repeat binding protein 4 (VvCBF4) transcriptional factor enhances freezing tolerance in wine grape. Plant Biotechnol J 10(1):105–124
Tong Z, Hong B, Yang Y, Li Q, Ma N, Ma C, Gao J (2009) Overexpression of two chrysanthemum DgDREB1 group genes causing delayed flowering or dwarfism in Arabidopsis. Plant Mol Biol 71(1–2):115–129
Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16(2):123–132
Volker P, Owen J, Borralho N (1994) Genetic variances and covariances for frost tolerance in Eucalyptus globulus and E. nitens. Silvae Genet 43(5–6):366–372
Wang Q, Guan Y, Wu Y, Chen H, Chen F, Chu C (2008) Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol Biol 67(6):589–602
Wang Z, Liu J, Guo H, He X, Wu W, Du J, Zhang Z, An X (2014) Characterization of two highly similar CBF/DREB1-like genes, PhCBF4a and PhCBF4b, in Populus hopeiensis. Plant Physiol Biochem 83:107–116
Wang L, Gao J, Qin X, Shi X, Luo L, Zhang G, Yu H, Li C, Hu M, Liu Q, Xu Y, Chen F (2015) JcCBF2 gene from Jatropha curcas improves freezing tolerance of Arabidopsis thaliana during the early stage of stress. Mol Biol Rep 42(5):937–945
Weigel D, Glazebrook J (2002) Arabidopsis: A laboratory manual. Cold Spring Harbor Laboratory Press, NY
Welling A, Palva E (2008) Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiol 147(3):1199–1211
Wisniewski M, Norelli J, Bassett C, Artlip T, Macarisin D (2011) Ectopic expression of a novel peach (Prunus persica) CBF transcription factor in apple (Malus × domestica) results in short-day induced dormancy and increased cold hardiness. Planta 233(5):971–983
Wisniewski M, Nassuth A, Teulières C, Marque C, Rowland J, Cao P, Brown A (2014) Genomics of cold hardiness in woody plants. Crit Rev Plant Sci 33(2–3):92–124
Xiao H, Siddiqua M, Braybrook S, Nassuth A (2006) Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant Cell Env 29(7):1410–1421
Xiao H, Tattersall E, Siddiqua M, Cramer G, Nassuth A (2008) CBF4 is a unique member of the CBF transcription factor family of Vitis vinifera and Vitis riparia. Plant Cell Env 31(1):1–10
Xu M, Li L, Fan Y, Wan J, Wang L (2011) ZmCBF3 overexpression improves tolerance to abiotic stress in transgenic rice (Oryza sativa) without yield penalty. Plant Cell Rep 30(10):1949–1957
Xue Y, Wang Y, Peng R, Zhen J, Zhu B, Gao J, Zhao W, Han H, Yao Q (2014) Transcription factor MdCBF1 gene increases freezing stress tolerance in transgenic Arabidopsis thaliana. Biol Plant 58(3):499–506
Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt Stress. Plant Cell 6(2):251–264
Zhang X, Fowler S, Cheng H, Lou Y, Rhee S, Stockinger E, Thomashow M (2004) Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J 39(6):905–919
Zhou M, Shen C, Wu L, Tang K, Lin J (2011) CBF-dependent signaling pathway: A key responder to low temperature stress in plants. Crit Rev Biotechnol 31(2):186–192
Zhou M, Xu M, Wu L, Shen C, Ma H, Lin J (2014) CbCBF from Capsella bursa-pastoris enhances cold tolerance and restrains growth in Nicotiana tabacum by antagonizing with gibberellin and affecting cell cycle signaling. Plant Mol Biol 85(3):259–275
Zhou M, Chen H, Wei D, Ma H, Lin J (2017) Arabidopsis CBF3 and DELLAs positively regulate each other in response to low temperature. Sci Rep 7:39819
Zhuang L, Yuan X, Chen Y, Xu B, Yang Z, Huang B (2015) PpCBF3 from cold-tolerant Kentucky Bluegrass involved in freezing tolerance associated with up-regulation of cold-related genes in transgenic Arabidopsis thaliana. PLoS One 10(7):e0132928
Acknowledgements
This work was funded by CORFO 12FBCT 16466 and Genómica Forestal S.A. A CONICYT Ph. D. scholarship to DN-C. Bioforest S.A. provided the E. globulus plant material used in this study. The technical assistance of Ms. Valeria Neira for sample handling and physiological data acquisition is greatly appreciated.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by F. Canovas.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Navarrete-Campos, D., Le Feuvre, R., Balocchi, C. et al. Overexpression of three novel CBF transcription factors from Eucalyptus globulus improves cold tolerance on transgenic Arabidopsis thaliana . Trees 31, 1041–1055 (2017). https://doi.org/10.1007/s00468-017-1529-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-017-1529-3