Abstract
Key message
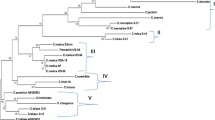
This study showed that Themachhi is genetically very close to C. medica and consistently grouped with the typical citron in all the phylogenetic trees; hence revealed the identity of ‘Themachhi’ as a natural wild type of C. medica.
Abstract
Taxonomic identity and well-supported phylogenetic relationships of doubtful and taxonomically uncertain taxa are highly requisite for developing crop improvement programmes and suitable conservation strategies. In Citrus, several natural varieties and hybrids of ambiguous origin have been existing due to several factors such as frequent intergeneric and interspecific hybridization, apomixis, bud mutations, polyploidy and diverse eco-geographical distribution. In Northeast India, an important unidentified type of Citrus, i.e. ‘Themachhi’ is found naturally in Garo Hills, Meghalaya, which is morphologically very close to C. medica and C. limon. The sequences of chloroplast non-coding region psbM-trnD of 16 accessions of Citrus were utilized to confirm the taxonomic identity of ‘Themachhi’ and also to infer its phylogenetic affinities to related taxa. The nucleotide frequencies were 32.86 % for A and T and 17.4 % for C and G. Pairwise genetic divergence between the accessions of Citrus ranged from 0.002 to 0.036 with an average of 0.017. ‘Themachhi’ showed least genetic divergence with C. medica (0.013) and highest divergence with C. limon (average 0.024). All the accessions of Citrus were grouped into three main clusters (C. medica, C. reticulata and C. maxima clusters) in both the MP and ML trees with high bootstrap value (>60 %). In the MP and ML trees, both accessions of ‘Themachhi’ were closely attached with C. medica cluster with high bootstrap value (98 %). Close genetic affinity and consistently grouping of “Themachhi” with typical citron (C. medica) in both the phylogenetic trees revealed the identity of ‘Themachhi’ as a natural wild type of C. medica.




Similar content being viewed by others
References
Abkenar AA, Isshiki S, Tashino Y (2004) Phylogenetic relationships in the true Citrus fruit trees revealed by PCR-RFLP analysis of cp DNA. Sci Hortic 102:233–242
Altschul SF, Thomas LM, Alejandro AS, Jinghui Z, Zheng Z, Webb M, David JL (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Anonymous (2010) Indian Horticulture Database, National Horticulture Board, Ministry of Agriculture, Gurgaon, Haryana
Araujo EF, Queiroz LP, Machado MA (2003) What is Citrus? Taxonomic implications from a study of cp-DNA evolution in the tribe Citreae (Rutaceae subfamily Aurantioideae). Org Divers Evol 3:55–62
Arora RK (2000) In Situ Conservation of Biological Diversities in Citrus. In: Proceedings of the Global Citrus Germplasm Network. Appendix 7, GCGN Meeting, 7–8 Dec
Barkley NA, Roose ML, Krueger RR, Federici CT (2006) Assessing genetic diversity and population structure in a Citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theor Appl Genet 112:1519–1531
Barrett HC, Rhodes AM (1976) A numerical taxonomic study of affinity relationships in cultivated Citrus and its close relatives. Syst Bot 1:105–136
Bayer RJ, Mabberley DJ, Morton CM, Cathy H, Sharma IK, Pfeil BE, Rich S, Hitchcock R, Sykes S (2009) A molecular phylogeny of the orange subfamily (Rutaceae: Aurantioideae) using nine cpDNA sequences. Am J Bot 96:668–685
Bhattacharya SC, Dutta S (1956) Classification of Citrus fruits of Assam, Sc Monogr 20, ICAR, New Delhi, p 110
De Candolle A (1886/1912) Origine des plantes cultivees (Origin of cultivated plants). 5th edn, Paris
Dugo G, Di Giacomo A (2002) Citrus: the genus Citrus, medicinal and aromatic plants—industrial profiles. Taylor & Francis group, London
Federici CT, Fang DQ, Scora RW, Roose ML (1998) Phylogenetic relationship within the genus Citrus (Rutaceae) and related genera as revealed by RFLP and RAPD analysis. Theor Appl Genet 96:812–822
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Herrero R, Asins MJ, Carbonell EA, Navarro L (1996) Genetic diversity in the orange subfamily Aurantioideae I. Intraspecies and Intragenus genetic variability. Theor Appl Genet 92:599–609
Jena SN, Kumar S, Nair KN (2009) Molecular phylogeny in Indian Citrus L. (Rutaceae) inferred through PCR-RFLP and trnL-trnF sequence data of chloroplast DNA. Sci Hortic 199:403–416
Kelchner SA (2000) The evolution of non-coding chloroplast DNA and its application in plant systematics. Ann Mo Bot Gard 87:499–527
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kumar S, Jena SN, Nair KN (2010) ISSR polymorphism in Indian Wild Orange, C. indica Tanaka (Rutaceae) and related wild species in North-east India. Sci Hortic 123:350–359
Kumar S, Nair KN, Jena SN (2012) Molecular differentiation in Indian Citrus L. (Rutaceae) inferred from nrDNA ITS sequence analysis. Genet Resour Crop Evol. doi:10.1007/s10722-012-9814-x
Kyndt T, Dung TN, Goetghebeur P, Toan HT, Gheysen G (2010) Analysis of ITS of the rDNA to infer phylogenetic relationship among Veitnamese Citrus accessions. Genet Res Crop Evol 57:183–192
Li J, Yue J, Shoup S (2006) Phylogenetics Of Acer (Aceroideae, Sapindaceae) based on nucleotide sequences of two chloroplast non-coding regions. Harv Papers Bot 11:101–115
Liang G, Xiong G, Guo Q, He Q, Li X (2007) AFLP analysis and the taxonomy of Citrus. Acta Hortic (ISHS) 760:137–142
Mabberley DJ (2004) Citrus (Rutaceae): a review of recent advances in etymology, systematics and medical applications. Blumea 49:481–498
Malik SK, Kumar S, Singh IP, Dhariwal OP, Chaudhury R (2013) Socio-economic importance, domestication trends and in situ conservation of wild Citrus species of Northeast India. Genet Res Crop Evol 60:1655–1671
Moore GA (2001) Oranges and lemons: clues to the taxonomy of Citrus from molecular markers. Trends Genet 17:536–540
Nair KN, Nayar MP (1997) Rutaceae. In: Hajra PK, Nair VJ, Daniel P (eds), Flora of India. Vol. IV, Botanical Survey of India, Calcutta, India, pp 229–407
Nicolosi E, Deng ZN, Genetile A, La Malfa S, Ciontinella G, Tribulata E (2000) Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor Appl Genet 100:1155–1166
Pang XM, Hu CG, Deng XX (2007) Phylogenetic relationships within Citrus and its related genera as inferred from AFLP markers. Genet Resour Crop Evol 54:429–436
Pessinaa D, Gentili R, Barcaccia G, Nicolè S, Rossic G, Barbestia S, Sgorbatia S (2011) DNA content, morphometric and molecular marker analyses of Citrus limonimedica, C. limon and C. medica for the determination of their variability and genetic relationships within the genus Citrus. Sci Horti 129:663–673
Ray BK, Deka PC (1999) Characterization of citrons growing in Northeast India using Hierarchical cluster analysis. Ind J Pl Genet Resour 12:50–55
Rogstad SH (1993) Saturated NaCl-CTAB solution as a means of field preservation leaves for DNA analysis. Taxon 41:701–708
Scora RW (1975) On the history and origin of Citrus. Bull Torrey Bot Club 102:369–375
Shahsavar AR, Izadpanah K, Tafazoli E, Tabatabaei BES (2007) Characterization of Citrus germplasm including unknown variants by inter-simple sequence repeat (ISSR) markers. Sci Hortic 112:310–314
Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 92:142–166
Swingle WT (1943) The botany of Citrus and its wild relatives. In: Webber HJ, Batchelor DL (eds) The Citrus industry, vol 1. University of California, Berkeley, pp 128–474
Swingle WT, Reece PC (1967) The botany of Citrus and orange relatives in the orange subfamily. In: Reuther W, Webber HJ, Batchelor DL (eds) The Citrus industry, vol 1. California Uni. Press, Berkeley, pp 190–340
Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol Biol Evol 9:678–687
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tanaka T (1954) Species problem in Citrus. Japanese Society for promotion of Science, Ueno, Tokyo, pp 152
Tanaka T (1977) Fundamental discussion of Citrus classification. Stud Citrol 14:1–6
Xu CJ, Bau L, Zhang B, Bei ZM, Ye XY, Zhang SL, Chen KS (2006) Parentage analysis of huyou (Citrus changshanensis) based on internal transcribed spacer sequences. Pl Breed 125:519–522
Acknowledgments
Authors are grateful to Director, NBPGR, New Delhi for encouragement and support. Thanks are also due to officers and staff of forest departments and ICAR institutes/centres in Northeast India for assistance and support during exploration and survey trips. Grant from NAIP-World Bank is greatly acknowledged for the Germplasm collection for this study. S.K. thanks Department of Biotechnology, Govt. of India for awarding Post Doctorate Fellowship.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Alia.
Rights and permissions
About this article
Cite this article
Kumar, S., Malik, S.K., Uchoi, A. et al. A new wild type of Citron (Citrus medica L., Rutaceae) identified through morphology and psbM-trnD spacer region of chloroplast DNA. Trees 28, 1115–1124 (2014). https://doi.org/10.1007/s00468-014-1022-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-014-1022-1