Abstract
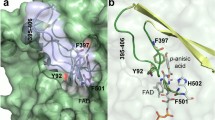
Arylacetonitrilases have been widely acknowledged as important alternatives to chemical catalysts for synthesizing optically pure 2-hydroxyphenylacetic acids from nitriles. In this work, two residues (Thr132 and Ser190) located at the catalytic tunnel in the active site of an arylacetonitrilase nitA from uncultured organisms were mutated separately by site-directed mutagenesis. Ser190 was demonstrated to be the critical position which has a greater influence on arylacetonitrilase nitA activity than Thr132. The replacement of serine at position 190 with glycine increases its activity toward mandelonitrile and (o, m, p)-chloromandelonitrile, whereas replacing it with leucine abolished its activity. The best mutant S190G exhibited threefold higher specific activity toward mandelonitrile compared with that of wild-type nitA, which rendered it promising for industrial application. Homology modeling and molecular docking experiments were in agreement with the kinetic assays and support the improved catalytic performance.







Similar content being viewed by others
References
Banerjee A, Kaul P, Banerjee UC (2006) Purification and characterization of an enantioselective arylacetonitrilase from Pseudomonas putida. Arch Microbiol 184:407–418
Groger H (2001) Enzymatic routes to enantiomerically pure aromatic alpha-hydroxy carboxylic acids: a further example for the diversity of biocatalysis. Adv Synth Catal 343:547–558
Xue YP, Yang YK, Lv SZ, Liu ZQ, Zheng YG (2016) High-throughput screening methods for nitrilases. Appl Microbiol Biotechnol 100:3421–3432
Martinkova L, Kren V (2010) Biotransformations with nitrilases. Curr Opin Chem Biol 14:130–137
Gong JS, Lu ZM, Li H, Shi JS, Zhou ZM, Xu ZH (2012) Nitrilases in nitrile biocatalysis: recent progress and forthcoming research. Microb Cell Fact 11:142
Gong JS, Li H, Lu ZM, Shi JS, Xu ZH (2015) Recent progress in the application of nitrilase in the biocatalytic synthesis of pharmaceutical intermediates. Prog Chem 27:448–458
Zhang XH, Liu ZQ, Xue YP, Xu M, Zheng YG (2016) Nitrilase-catalyzed conversion of (R, S)-mandelonitrile by immobilized recombinant Escherichia coli cells harboring nitrilase. Biotechnol Appl Biochem 63:479–489
Singh R, Sharma R, Tewari N, Geetanjali Rawat DS (2006) Nitrilase and its application as a ‘green’ catalyst. Chem Biodivers 3:1279–1287
Yamamoto K, Fujimatsu I, Komatsu K (1992) Purification and characterization of the nitrilase from Alcaligenes faecalis ATCC8750 responsible for enantioselective hydrolysis of mandelonitrile. J Ferment Bioeng 73:425–430
Rustler S, Muller A, Windeisen V, Chmura A, Fernandes BCM, Kiziak C, Stolz A (2007) Conversion of mandelonitrile and phenylglycinenitrile by recombinant E. coli cells synthesizing a nitrilase from Pseudomonas fluorescens EBC191. Enzyme Microb Technol 40:598–606
Wang HL, Sun HH, Wei DZ (2013) Discovery and characterization of a highly efficient enantioselective mandelonitrile hydrolase from Burkholderia cenocepacia J2315 by phylogeny-based enzymatic substrate specificity prediction. BMC Biotechnol 13:14
Zhang CS, Zhang ZJ, Li CX, Yu HL, Zheng GW, Xu JH (2012) Efficient production of (R)-o-chloromandelic acid by deracemization of o-chloromandelonitrile with a new nitrilase mined from Labrenzia aggregata. Appl Microbiol Biotechnol 95:91–99
Sun HH, Gao WY, Fan HY, Wang HL, Wei DZ (2015) Cloning, purification and evaluation of the enzymatic properties of a novel arylacetonitrilase from Luminiphilus syltensis NOR5-1B: a potential biocatalyst for the synthesis of mandelic acid and its derivatives. Biotechnol Lett 37:1655–1661
He YC, Xu JH, Xu Y, Ouyang LM, Pan J (2007) Biocatalytic synthesis of (R)-(−)-mandelic acid from racemic mandelonitrile by a newly isolated nitrilase-producer Alcaligenes sp. ECU0401. Chin Chem Lett 18:677–680
Nagasawa T, Mauger J, Yamada H (1990) A novel nitrilase, arylacetonitrilase, of Alcaligenes faecalis JM3. Purification and characterization. Eur J Biochem 194:765–772
Layh N, Parratt J, Willetts A (1998) Characterization and partial purification of an enantioselective arylacetonitrilase from Pseudomonas fluorescens DSM 7155. J Mol Catal B Enzym 5:467–474
Banerjee A, Dubey S, Kaul P, Barse B, Piotrowski M, Banerjee U (2009) Enantioselective nitrilase from Pseudomonas putida: cloning, heterologous expression, and bioreactor studies. Mol Biotechnol 41:35–41
Liu ZQ, Dong LZ, Cheng F, Xue YP, Wang YS, Ding JN, Zheng YG, Shen YC (2011) Gene cloning, expression, and characterization of a nitrilase from Alcaligenes faecalis ZJUTB10. J Agric Food Chem 59:11560–11570
Sun HH, Wang HL, Gao WY, Chen LF, Wu K, Wei DZ (2015) Directed evolution of nitrilase PpL19 from Pseudomonas psychrotolerans L19 and identification of enantiocomplementary mutants toward mandelonitrile. Biochem Biophys Res Commun 468:820–825
Vesela AB, Petrickova A, Weyrauch P, Martinkova L (2013) Heterologous expression, purification and characterization of arylacetonitrilases from Nectria haematococca and Arthroderma benhamiae. Biocatal Biotransform 31:49–56
Vesela AB, Krenkova A, Martinkova L (2015) Exploring the potential of fungal arylacetonitrilases in mandelic acid synthesis. Mol Biotechnol 57:466–474
Robertson DE, Chaplin JA, DeSantis G, Podar M, Madden M, Chi E, Richardson T, Milan A, Miller M, Weiner DP, Wong K, McQuaid J, Farwell B, Preston LA, Tan X, Snead MA, Keller M, Mathur E, Kretz PL, Burk MJ, Short JM (2004) Exploring nitrilase sequence space for enantioselective catalysis. Appl Environ Microbiol 70:2429–2436
Xue YP, Shi CC, Xu Z, Jiao B, Liu ZQ, Huang JF, Zheng YG, Shen YC (2015) Design of nitrilases with superior activity and enantioselectivity towards sterically hindered nitrile by protein engineering. Adv Synth Catal 357:1741–1750
Schreiner U, Hecher B, Obrowsky S, Waich K, Klempier N, Steinkellner G, Gruber K, Rozzell JD, Glieder A, Winkler M (2010) Directed evolution of Alcaligenes faecalis nitrilase. Enzyme Microb Technol 47:140–146
Liu ZQ, Zhang XH, Xue YP, Xu M, Zheng YG (2014) Improvement of Alcaligenes faecalis nitrilase by gene site saturation mutagenesis and its application in stereospecific biosynthesis of (R)-(−)-mandelic acid. J Agric Food Chem 62:4685–4694
Wang HL, Gao WY, Sun HH, Chen LF, Zhang LJ, Wang XD, Wei DZ (2015) Protein engineering of a nitrilase from Burkholderia cenocepacia J2315 for efficient and enantioselective production of (R)-o-chloromandelic acid. Appl Environ Microbiol 81:8469–8477
Vergne-Vaxelaire C, Bordier F, Fossey A, Besnard-Gonnet M, Debard A, Mariage A, Pellouin V, Perret A, Petit JL, Stam M, Salanoubat M, Weissenbach J, De Berardinis V, Zaparucha A (2013) Nitrilase activity screening on structurally diverse substrates: providing biocatalytic tools for organic synthesis. Adv Synth Catal 355:1763–1779
Chen CS, Fujimoto Y, Girdaukas G, Sih CJ (1982) Quantitative analyses of biochemical kinetic resolutions of enantiomers. J Am Chem Soc 104:7294–7299
Kaul P, Stolz A, Banerjee UC (2007) Cross-linked amorphous nitrilase aggregates for enantioselective nitrile hydrolysis. Adv Synth Catal 349:2167–2176
O’Reilly C, Turner PD (2003) The nitrilase family of CN hydrolysing enzymes—a comparative study. J Appl Microbiol 95:1161–1174
Babbitt PC, Gerlt JA (1997) Understanding enzyme superfamilies—chemistry as the fundamental determinant in the evolution of new catalytic activities. J Biol Chem 272:30591–30594
Nakai T, Hasegawa T, Yamashita E, Yamamoto M, Kumasaka T, Ueki T, Nanba H, Ikenaka Y, Takahashi S, Sato M, Tsukihara T (2000) Crystal structure of N-carbamyl-d-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases. Structure 8:729–737
Williamson DS, Dent KC, Weber BW, Varsani A, Frederick J, Thuku RN, Cameron RA, van Heerden JH, Cowan DA, Sewell BT (2010) Structural and biochemical characterization of a nitrilase from the thermophilic bacterium, Geobacillus pallidus RAPc8. Appl Microbiol Biotechnol 88:143–153
Watanabe A, Yano K, Ikebukuro K, Karube I (1998) Investigation of the potential active site of a cyanide dihydratase using site-directed mutagenesis. BBA Protein Struct M 1382:1–4
Bhatia SK, Mehta PK, Bhatia RK, Bhalla TC (2014) Optimization of arylacetonitrilase production from Alcaligenes sp. MTCC 10675 and its application in mandelic acid synthesis. Appl Microbiol Biotechnol 98:83–94
Zhang ZJ, Xu JH, He YC, Ouyang LM, Liu YY (2011) Cloning and biochemical properties of a highly thermostable and enantioselective nitrilase from Alcaligenes sp. ECU0401 and its potential for (R)-(−)-mandelic acid production. Bioprocess Biosyst Eng 34:315–322
Acknowledgements
This work was supported by the NSFC (No. 21676254).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that there is no conflict of interest.
Rights and permissions
About this article
Cite this article
Xue, YP., Jiao, B., Hua, DE. et al. Improving catalytic performance of an arylacetonitrilase by semirational engineering. Bioprocess Biosyst Eng 40, 1565–1572 (2017). https://doi.org/10.1007/s00449-017-1812-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-017-1812-0