Abstract
Main conclusion
This study is the first endeavor on mining of miRNAs and analyzing their involvement in development and secondary metabolism of an endangered medicinal herb Picrorhiza kurroa (P. kurroa ).
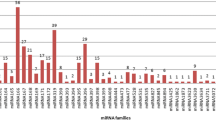
miRNAs are ubiquitous non-coding RNA species that target complementary sequences of mRNA and result in either translational repression or target degradation in eukaryotes. The role of miRNAs has not been investigated in P. kurroa which is a medicinal herb of industrial value due to the presence of secondary metabolites, picroside-I and picroside-II. Computational identification of miRNAs was done in 6 transcriptomes of P. kurroa generated from root, shoot, and stolon organs varying for growth, development, and culture conditions. All available plant miRNA entries were retrieved from miRBase and used as backend datasets to computationally identify conserved miRNAs in transcriptome data sets. Total 18 conserved miRNAs were detected in P. kurroa followed by target prediction and functional annotation which suggested their possible role in controlling various biological processes. Validation of miRNA and expression analysis by qRT-PCR and 5′ RACE revealed that miRNA-4995 has a regulatory role in terpenoid biosynthesis ultimately affecting the production of picroside-I. miR-5532 and miR-5368 had negligible expression in field-grown samples as compared to in vitro-cultured samples suggesting their role in regulating P. kurroa growth in culture conditions. The study has thus identified novel functions for existing miRNAs which can be further validated for their potential regulatory role.






Similar content being viewed by others
Abbreviations
- miRNA:
-
MicroRNA
- P. kurroa :
-
Picrorhiza kurroa
- PKS-25:
-
P. kurroa in vitro-cultured shoots grown at 25 °C
- PKS-15:
-
P. kurroa in vitro-cultured shoots grown at 15 °C
- PKR-25:
-
P. kurroa in vitro-cultured roots grown at 25 °C
- PKSS:
-
P. kurroa field-grown shoots
- PKSR:
-
P. kurroa field-grown roots
- PKSTS:
-
P. kurroa field-grown stolons
- P-I and II:
-
Picroside I and II
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bartel B, Bartel DP (2003) MicroRNAs: at the root of plant development? Plant Physiol 132:709–717
Cassells AC, Curry RF (2001) Oxidative stress and physiological, epigenetic and genetic variability in plant tissue culture: implications for micropropagators and genetic engineers. Plant Cell Tiss Org 64:145–157
Chopra RN, Ghosh S (1934) Some common indigenous remedies. Indian J Med Res 22:263–264
Das A, Mondal TK (2010) Computational identification of conserved microRNAs and their targets in tea (Camellia sinensis). Am J Plant Sci 1:77
Davuluri GR, van Tuinen A, Fraser PD, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J (2005) Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat Biotechnol 23:890–895
Dhawan B (1995) Picroliv—a new hepatoprotective agent from an Indian medicinal plant, Picrorhiza kurroa. Med Chem Res 5:595–605
Dudareva N, Pichersky E, Gershenzon J (2004) Biochemistry of plant volatiles. Plant Physiol 135:1893–1902
Fu C, Sunkar R, Zhou C, Shen H, Zhang JY, Matts J, Wolf J, Mann DGJ, Stewart CN Jr, Tang Y, Wang ZY (2012) Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol J 10:443–452
Gahlan P, Singh HR, Shankar R, Sharma N, Kumari A, Chawla V, Ahuja PS, Kumar S (2012) De novo sequencing and characterization of Picrorhiza kurrooa transcriptome at two temperatures showed major transcriptome adjustments. BMC Genom 13:126
German MA, Luo S, Schroth G, Meyers BC, Green PJ (2009) Construction of parallel analysis of RNA ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat Protoc 4:356–362
Gou J, Felippes F, Liu C, Weigel D, Wang J (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23:1512–1522
Gupta N, Sharma SK, Rana JC, Chauhan RS (2011) Expression of flavonoid biosynthesis genes vis-a-vis rutin content variation in different growth stages of Fagopyrum species. J Plant Physiol 168:2117–2123
He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q, Li J (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42:541–544
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Kawoosa T, Singh H, Kumar A, Sharma SK, Devi K, Dutt S (2010) Light and temperature regulated terpene biosynthesis: hepatoprotective monoterpene picroside accumulation in Picrorhiza kurrooa. Funct Integr Genomic 10:393–404
Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N (2005) Combinatorial microRNA target predictions. Nat Genet 37(5):495–500
Kumar V, Sood H, Sharma M, Chauhan RS (2012) A proposed biosynthetic pathway of picrosides linked through the detection of biochemical intermediates in the endangered medicinal herb Picrorhiza kurroa. Phytochem Anal 24:598–602
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Legrand S, Valot N, Nicole F, Moja S, Baudino S, Jullien F, Magnard JL, Caissard JC, Legendre L (2010) One-step identification of conserved miRNAs, their targets, potential transcription factors and effector genes of complete secondary metabolism pathways after 454 pyrosequencing of calyx cDNAs from the Labiate Salvia sclarea L. Gene 450:55–62
Li H, Zhao X, Dai H, Wu W, Mao W, Zhang Z (2012) Tissue culture responsive microRNAs in strawberry. Plant Mol Biol Rep 30:1047–1054
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769–773
Mahmoud SS, Croteau RB (2002) Strategies for transgenic manipulation of monoterpene biosynthesis in plants. Trends Plant Sci 7:366–373
Mallory AC, Vaucheret H (2006) Functions of microRNAs and related small RNAs in plants. Nat Genet 38:S31–S36
Martin A, Adam H, Díaz-Mendoza M, Zurczak M, González-Schain ND, Suárez-López P (2009) Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 136:2873–2881
Mehra TS, Chand R, Sharma Y (2007) Reproductive biology of Picrorhiza kurroa—a critically endangered high value temperate medicinal plant. Open Acc J Med Arom Pl 1:40–43
Ng DWK, Zhang C, Miller M, Palmer G, Whiteley M, Tholl D, Chen ZJ (2011) cis-and trans-Regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids. Plant Cell 23:1729–1740
Pandit S, Shitiz K, Sood H, Chauhan RS (2012) Differential biosynthesis and accumulation of picrosides in an endangered medicinal herb Picrorhiza kurroa. J Plant Biochem Biot 22:335–342
Pandit S, Shitiz K, Sood H, Naik PK, Chauhan RS (2013) Expression pattern of fifteen genes of non-mevalonate (MEP) and mevalonate (MVA) pathways in different tissues of endangered medicinal herb Picrorhiza kurroa with respect to picrosides content. Mol Biol Rep 40:1053–1063
Pradervand S, Weber J, Lemoine F, Consales F, Paillusson A, Dupasquier M, Thomas J, Richter H, Kaessmann H, Beaudoing E (2010) Concordance among digital gene expression, microarrays, and qPCR when measuring differential expression of microRNAs. Biotechniques 48:219–222
Qiao M, Zhao Z, Song Y, Liu Z, Cao L, Yu Y, Li S, Xiang F (2012) Proper regeneration from in vitro cultured Arabidopsis thaliana requires the microRNA-directed action of an auxin response factor. Plant J 71:14–22
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Gene Dev 16:1616–1626
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110:513–520
Robert-Seilaniantz A, MacLean D, Jikumaru Y, Hill L, Yamaguchi S, Kamiya Y, Jones JDG (2011) The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J 67:218–231
Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF (2010) Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137:103–112
Samanani N, Park SU, Facchini PJ (2005) Cell type specific localization of transcripts encoding nine consecutive enzymes involved in protoberberine alkaloid biosynthesis. Plant Cell 17:915–926
Sato F, Tsuchiya S, Meltzer SJ, Shimizu K (2011) MicroRNAs and epigenetics. FEBS J 278:1598–1609
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Shamimuzzaman M, Vodkin L (2012) Identification of soybean seed developmental stage-specific and tissue-specific miRNA targets by degradome sequencing. BMC Genom 13:310
Singh TR, Gupta A, Suravajhala P (2013) Challenges in the miRNA research. Int J Bioinformatics Res 9:576–583
Smit HF (2000) Picrorhiza scrophulariiflora, from traditional use to immunomodulatory activity. Dissertation, Proefschrift, Universiteit Utrecht, The Netherlands
Sood H, Chauhan RS (2010) Biosynthesis and accumulation of a medicinal compound, picroside-I, in cultures of Picrorhiza kurroa Royle ex Benth. Plant Cell Tiss Org 100:113–117
Stuppner H, Wagner H (1989) New cucurbitacin glycosides from Picrorhiza kurroa. Planta Med 55:559–563
Sud A, Chauhan RS, Tandon C (2013) Identification of imperative enzymes by differential protein expression in Picrorhiza kurroa under metabolite accumulating and non-accumulating conditions. Protein Peptide Lett 20:826–835
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687
Wang CM, Liu P, Sun F, Li L, Liu P, Ye J, Yue GH (2012) Isolation and Identification of miRNAs in Jatropha curcas. Int J Biol Sci 8(3):418–429
Wu G, Park MY, Conway SR, Wang J-W, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138:750–759
Xie F, Frazier TP, Zhang B (2010) Identification and characterization of microRNA and their targets in the bioenergy plant switchgrass (Panicum virgatum). Planta 232:417–434
Xie K, Shen J, Hou X, Yao J, Li X, Xiao J, Xiong L (2012) Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiol 158:1382–1394
Xu W, Cui Q, Li F, Liu A (2013) Transcriptome-wide identification and characterization of microRNAs from castor bean (Ricinus communis L.). PLoS ONE 8:e69995
Zhang B, Pan X, Anderson TA (2006) Identification of 188 conserved maize microRNAs and their targets. FEBS Lett 580:3753–3762
Zhou M, Lou H (2013) MicroRNA-mediated gene regulation: potential applications for plant genetic engineering. Plant Mol Biol 83:59–75
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Acknowledgments
The authors are thankful to the Department of Biotechnology, Ministry of Science & Technology, Government of India, for providing research grant in the form of a program support on high value medicinal plants to RSC.
Conflict of interest
Authors declare that they do not have any conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vashisht, I., Mishra, P., Pal, T. et al. Mining NGS transcriptomes for miRNAs and dissecting their role in regulating growth, development, and secondary metabolites production in different organs of a medicinal herb, Picrorhiza kurroa . Planta 241, 1255–1268 (2015). https://doi.org/10.1007/s00425-015-2255-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-015-2255-y