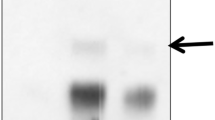
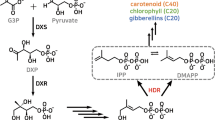
Abstract
Picrorhiza (Picrorhiza kurrooa) is an endangered medicinal plant with well-known hepatoprotective activity attributed to monoterpenoid picrosides. The present article details on regulatory genes of terpenoid metabolism, 3-hydroxy-3-methylglutaryl coenzyme A reductase (pkhmgr) and 1-deoxy-D-xylulose-5-phosphate synthase (pkdxs) from picrorhiza. Since no molecular information was available, these genes were cloned to full-length by degenerate primers and rapid amplification of cDNA ends, followed by cloning of the upstream sequences that showed the presence of core sequences for light and temperature responsiveness. Electrophoretic mobility shift assay confirmed binding of protein to these motifs. Expression of pkhmgr and pkdxs was up-regulated at 15°C as compared to at 25°C as well as under light as compared to dark conditions. Picrosides content exhibited the trend similar to gene expression. To rule out the possible limitation of carbon pool under dark condition, plantlets of picrorhiza were raised in vitro in Murashige and Skoog medium supplemented with 3% sucrose. Results showed similar up-regulation of both the genes and the higher picrosides content in in vitro raised plantlets in the presence of light. Data suggested the important roles played by light and temperature in regulating pkhmgr and pkdxs, and the picrosides level in picrorhiza.





Similar content being viewed by others
Abbreviations
- aa:
-
amino acid
- DXS:
-
1-deoxy-D-xylulose-5-phosphate synthase
- EMSA:
-
electrophoretic mobility shift assay
- GDP:
-
geranyl diphosphate
- HMGR:
-
3-hydroxy-3-methylglutaryl coenzyme A reductase
- MEP:
-
methylerythritol phosphate
- MVA:
-
mevalonate
- PLACE:
-
plant cis-acting regulatory elements
References
Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 28:37636–37645
Aoyagi K, Beyou A, Moon K, Fang L, Ulrich T (1993) Isolation and characterization of cDNAs encoding wheat 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Physiol 102:623–628
Busk PK, Pages M (1997) Microextraction of nuclear proteins from single maize embryos. Plant Mol Biol Rep 15:371–376
Chahed K, Oudin A, Guivarc’h N, Hamdi S, Che’nieux J-H, Rideau M, Clastre M (2000) 1-Deoxy-d-xylulose 5-phosphate synthase from periwinkle: cDNA identification and induced gene expression in terpenoidindole alkaloid-producing cells. Plant Physiol Bioch 38:559–566
Chandra S (2004) Effect of altitude on energy exchange characteristics of some alpine medicinal crops from central Himalayas. J Agron Crop Sci 190:13–20
Chettri N, Sharma E, Lama SD (2005) Non-timber forest produces utilization, distribution and status in a trekking corridor of Sikkim, India. Lyonia 8:89–101
Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABRE/ACGT binding sites ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275:1723–1730
Cordoba E, Salmi M, León P (2009) Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J Exp Bot 60:2933–2943
Croteau RB, Davis EM, Ringer KL, Wildung MR (2005) (-)-Menthol biosynthesis and molecular genetics. Naturwissenschaften 92:562–577
Degenhardt J, Tobin EM (1996) A DNA binding activity for one of two closely defined phytochrome regulatory elements in an Lhcb promoter is more abundant in etiolated than in green plants. Plant Cell 8:31–41
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Dunn MA, White AJ, Vural S, Hughes MA (1998) Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.). Plant Mol Biol 38:551–564
Dutt S, Kiddle G, Singh B, Khambay B, Foyer CH (2004) Differential accumulation of picrosides in Picrorhiza kurrooa Royle ex Benth plants. http://www.rothamsted-international.org/publications_posters.shtml
Estevez JM, Cantero A, Reindl A, Reichler S, Leon P (2001) 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastid isoprenoid biosynthesis in plants. J Biol Chem 25:22901–22909
Evans T, Reitman M, Felsenfeld G (1988) An erythrocyte specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci USA 85:5976–5980
Fey V, Wagner R, Brautigam K, Wirtz M, Hell R, Dietzmann A, Leister D, Oelmüller R, Pfannschmidt T (2005) Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J Biol Chem 280:5318–5328
Floersheim GL, Bieri A, Koenig R, Pletscher A (1990) Protection against Amanita phalloides by the iridoid glycoside mixture of Picrorhiza kurrooa (kutkin). Agents Actions 29:386–387
Fuglevand G, Jackson JA, Jenkins GI (1996) UV-B, UV-A, and blue light signal transduction pathways interact synergistically to regulate chalcone synthase gene expression in Arabidopsis. Plant Cell 8:2347–2357
Ghawana S, Singh K, Raizada J, Rani A, Bhardwaj PK, Kumar S (2007) A method for rapid isolation of RNA and kit thereof. WO 2007/113614
Gilmartin PM, Sarokin L, Memelink J, Chua NH (1990) Molecular light switches for plant genes. Plant Cell 2:369–378
Green PJ, Yong MH, Cuouo M, Kano-Murakami Y, Silverstein P, Chua NH (1988) Binding site requirements for the pea GT-1 correlate with sequences required for light dependent transcriptional activation of the rbcS-3A gene. EMBO J 7:4035–4044
Hampel D, Mosandl A, Wiist M (2005) Biosynthesis of mono- and sesquiterpenes in carrot roots and leaves (Daucus carota L.): metabolic cross talk of cytosolic mevalonate and plastidial methylerythritol phosphate pathways. Phytochemistry 66:305–311
Hampel D, Mosandl A, Wiist M (2006) Biosynthesis of mono- and sesquiterpenes in strawberry fruits and foliage: 2H labeling studies. J Agric Food Chem 54:1473–1478
Hannah MA, Heyer AG, Hincha DK (2005) A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet 1:e26s
Hemm MR, Rider SD, Ogas J, Murry DJ, Chapple C (2004) Light induces phenylpropanoid metabolism in Arabidopsis roots. Plant J 38:765–778
Hennighausen L, Lubon H (1985) Interaction of protein with DNA in vitro. Method Enzymol 152:721–735
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27:297–300
Hsieh MH, Goodman HM (2005) The arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis. Plant Physiol 138:641–653
Hudson ME, Quail PH (2003) Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol 133:1605–1616
Juan ME, Araceli C, Andreas R, Stuart R, Patricia L (2001) 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J Biol Chem 276:22901–22909
Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, Peacock WJ, Dolferus R, Dennis ES (2002) Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 14:2481–2494
Korth KL, Jaggard DAW, Dixon RA (2000) Developmental and light-regulated post-translational control of 3-hydroxy-3-methylglutaryl-CoA reductase levels in potato. Plant J 23:507–516
Lange BM, Severin K, Bechthold A, Heide L (1998) Regulatory role of microsomal 3-hydroxy-3-methylglutaryl-coenzyme A reductase for shikonin biosynthesis in Lithospermum erythrorhizon cell suspension cultures. Planta 204:234–241
Learned RM, Connolly EL (1997) Light modulates the spatial patterns of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene expression in Arabidopsis thaliana. Plant J 11:499–511
Leivar P, Gonzalez VM, Castel S, Trelease RN, Lesias C, Arro M, Boronat A, Campos N, Ferrer A, Busquets X (2005) Subcellular localization of arabidopsis 3-hydroxy-3-methylglutaryl-coenzyme a reductase. Plant Physiol 137:57–69
Lois LM, Rodriguez-Concepcion M, Gallego F, Campos N, Boronat A (2000) Carotenoid biosynthesis during tomato fruit development: regulatory role of 1-deoxy-D-xylulose 5-phosphate synthase. Plant J 22:503–513
Luper S (1998) A review of plants used in the treatment of liver disease: part 1. Altern Med Rev 3:410–421
Mahmoud SS, Croteau RB (2002) Strategies for transgenic manipulation of monoterpene biosynthesis in plants. Trends Plant Sci 7:366–373
Mosaleeyanon K, Zobayed SMA, Afreen F, Kozai T (2005) Relationships between net photosynthetic rate and secondary metabolite contents in St. John’s wort. Plant Sci 169:523–531
Munoz-Bertomeu J, Arrillaga I, Ros R, Segura J (2006) Up-regulation of 1-deoxy-D-xylulose-5-phosphate synthase enhances production of essential oils in transgenic spike lavender. Plant Physiol 142:890–900
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol Plant 15:473–497
Nayar MP, Sastri ARK (1990) Red data plants of India. CSIR Publication, New Delhi, p 271
Nogués I, Brilli F, Loreto F (2006) Dimethylallyl diphosphate and geranyl diphosphate pools of plant species characterized by different isoprenoid emissions. Plant Physiol 141:721–730
Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, Kim MC, Park CY, Jeong JC, Moon BC, Lee JH, Yoon HW, Lee SH, Chung WS, Lim CO, Lee SY, Hong JC, Cho MJ (2004) Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol 135:2150–2161
Reyes JC, Muro-Pastor MI, Florencio FJ (2004) The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol 134:1718–1732
Sacchettini JC, Poulter CD (1997) Creating isoprenoid diversity. Science 277:1788–1789
Singh K, Raizada J, Bhardwaj P, Ghawana S, Rani A, Singh H, Kaul K, Kumar S (2004) 26S rRNA-based internal control gene primer pair for reverse transcription-polymerase chain reaction-based quantitative expression studies in diverse plant species. Anal Biochem 335:330–333
Singh N, Gupta AP, Singh B, Kaul VK (2005) Quantification of picroside-I and picroside-II in Picrorhiza kurrooa by HPTLC. J Liq Chromatogr & Rel Technol 28:1679–1691
Soitamo AJ, Piippo M, Allahverdiyeva Y, Battchikova N, Aro EM (2008) Light has a specific role in modulating Arabidopsis gene expression at low temperature. BMC Plant Biol 8:13
Streb P, Shang W, Feierabend J, Bligny R (1998) Divergent strategies of photoprotection in high-mountain plants. Planta 207:313–324
Teakle GR, Manfield IW, Graham JF, Philip M, Gilmartin PM (2002) Arabidopsis thaliana GATA factors: organisation, expression and DNA-binding characteristics. Plant Mol Biol 50:43–57
Terzaghi WB, Cashmore AR (1995) Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol 46:445–474
Tjaden G, Edwards JW, Coruzzi GM (1995) Cis elements and trans-acting factors affecting regulation of a non-photosynthetic light-regulated gene for chloroplast glutamine synthetase. Plant Physiol 108:1109–1117
Udvardi MK, Kakar K, Wandrey M, Montanari O, Murray J, Andriankaja A, Zhang JY, Benedito V, Hofer JM, Chueng F, Town CD (2007) Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiol 144:538–549
Villain P, Mache R, Zhou DX (1996) The mechanism of GT element-mediated cell type-specific transcriptional control. J Biol Chem 271:32593–32598
Walter MH, Hans J, Strack D (2002) Two distantly related genes encoding 1-deoxy-d-xylulose 5-phosphate synthases: differential regulation in shoots and apocarotenoid-accumulating mycorrhizal roots. Plant J 31:243–254
Wang Y, Guo B, Zhang F, Yao H, Miao Z, Tang K (2007) Molecular cloning and functional analysis of the gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase from hazel (Corylus avellana L. Gasaway). J Biochem Mol Biol 40:861–869
Wise ML, Croteau R (1998) Comprehensive natural products chemistry: isoprenoid biosynthesis. In: Cane DE (ed) Pergamon, Oxford
Wong RJ, McCormack DK, Russell DW (1982) Plastid 3-hydroxy-3-methylglutaryl coenzyme A reductase has distinctive kinetic and regulatory features: properties of the enzyme and positive phytochrome control of activity in pea seedlings. Arch Biochem Biophys 216:631–638
Yang Z, Park H, Lacy GH, Cramer CL (1991) Differential activation of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes by wounding and pathogen challenge. Plant Cell 3:397–405
Acknowledgments
We acknowledge financial support from Council of Scientific and Industrial Research (CSIR) for work on gene cloning through Mission Mode program entitled “Development of medicinal plant chemotypes for enhanced marker and value added compounds (COR002)”. Financial support from the Department of Biotechnology (DBT), Govt. of India is duly acknowledged for funding the project entitled “Molecular cloning and characterization of regulatory genes involved in picrosides metabolism in Picrorhiza kurrooa” sanctioned vide sanction order number BT/PR6614/PBD/17/425/2005 to work on promoters. TK thanks CSIR, India, for awarding CSIR Diamond Jubilee Research Intern Award; HS thanks CSIR for awarding junior and senior research fellowships. AK, KD, and SKS gratefully acknowledge DBT for providing assistantship. Technical help provided by Mr. Digvijay Singh in gene sequencing is duly acknowledged. The manuscript represents IHBT communication number 0812.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Electronic Supplementary Table S1
Comparison of predicted secondary structures of HMGR and DXS of picrorhiza with those from other plants. Protein sequences were retrieved from the NCBI databank and secondary structural prediction was carried out using ExPASy server tool SOPMA (http://www.npsa-pbil.ibcp.fr/) by keeping similarity threshold 8 and conformational states 4. (DOC 74 kb)
Electronic Supplementary Fig. S1
Multiple sequence alignment of deduced amino acids encoded by pkhmgr (3-hydroxy-3-methylglutaryl coenzyme A reductase; S1 a) and pkdxs (1-deoxy-D-xylulose-5-phosphate synthase; S1 b) of P. kurrooa. Alignment was performed by Accelrys software tool (Accelrys Gene software, version 2.5 of Accelrys software Inc., USA). Sequences denoted are: P. kurrooa -PK (ABC74565), Nicotiana tabacum -NT (AAB87727), Eucommia ulmoides -EU (AAV54051), Andrographis paniculata -AP (AAP14352), Litchi chinensis -LC (ABF56181), Mentha x piperita -MP (AAC33513), Antirrhinum majus -AM (AAW28999), Tagetes erecta -TE (AAG10432), Catharanthus roseus -CR (CAA09804). Accession number of the deduced amino acid sequences is shown in the respective parenthesis. (PPT 394 kb)
Electronic Supplementary Fig. S2
Conserved domains of deduced proteins pkHMGR (3-hydroxy-3-methylglutaryl coenzyme A reductase) and pkDXS (1-deoxy-D-xylulose-5-phosphate synthase) of picrorhiza. Deduced amino acid sequences were analyzed for the location of conserved domains using Conserved Domain Database available at NCBI (http://www.ncbi.nlm.nih.gov/ Structure/cdd/wrpsb.cgi). (PPT 39 kb)
Electronic Supplementary Fig. S3
Predicted secondary structure of deduced proteins, pkHMGR (3-hydroxy-3-methylglutaryl coenzyme A reductase) and pkDXS (1-deoxy-D-xylulose-5-phosphate synthase) using ExPASy server tool, SOPMA (http://www.npsa-pbil.ibcp.fr/) by keeping similarity threshold 8 and conformational states 4. Helices, sheets, turns, and coils are indicated with the longest, the second longest, the second shortest and the shortest vertical lines, respectively. (PPT 104 kb)
Rights and permissions
About this article
Cite this article
Kawoosa, T., Singh, H., Kumar, A. et al. Light and temperature regulated terpene biosynthesis: hepatoprotective monoterpene picroside accumulation in Picrorhiza kurrooa . Funct Integr Genomics 10, 393–404 (2010). https://doi.org/10.1007/s10142-009-0152-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-009-0152-9