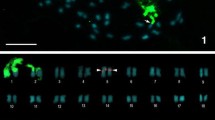
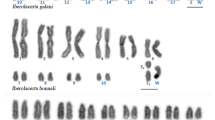
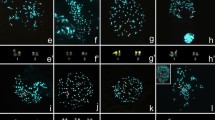
Abstract
Crocodilians have maintained very similar karyotype structures and diploid chromosome numbers for around 100 million years, with only minor variations in collinearity. Why this karyotype structure has largely stayed unaltered for so long is unclear. In this study, we analyzed the karyotypes of six species belonging to the genera Crocodylus and Osteolaemus (Crocodylidae, true crocodiles), among which the Congolian endemic O. osborni was included and investigated. We utilized various techniques (differential staining, fluorescence in situ hybridization with repetitive DNA and rDNA probes, whole chromosome painting, and comparative genomic hybridization) to better understand how crocodile chromosomes evolved. We studied representatives of three of the four main diploid chromosome numbers found in crocodiles (2n = 30/32/38). Our data provided new information about the species studied, including the identification of four major chromosomal rearrangements that occurred during the karyotype diversification process in crocodiles. These changes led to the current diploid chromosome numbers of 2n = 30 (fusion) and 2n = 38 (fissions), derived from the ancestral state of 2n = 32. The conserved cytogenetic tendency in crocodilians, where extant species keep near-ancestral state, contrasts with the more dynamic karyotype evolution seen in other major reptile groups.








Similar content being viewed by others
Data availability
All data presented here are included in the article and its Supplementary Information. Additional questions should be directed to the corresponding author.
References
Amavet P, Markariani R, Fenocchio A (2003) Comparative cytogenetic analysis of the South American alligators Caiman latirostris and Caiman yacare (Reptilia, Alligatoridae) from Argentina. Caryologia 56:489–493. https://doi.org/10.1080/00087114.2003.10589361
Ariyaraphong N, Wongloet W, Wattanadilokchatkun P, Panthum T, Singchat W, Thong T, Lisachov A, Ahmad SF, Muangmai N, Han K, Duengkae P, Temsiripong Y, Srikulnath K (2023) Should the identification guidelines for Siamese Crocodiles be revised? Differing post-occipital scute scale numbers show phenotypic variation does not result from hybridization with Saltwater Crocodiles. Biology 12:535. https://doi.org/10.3390/biology12040535
Axelrod DI (1952) A theory of Angiosperm evolution. Evolution 1:29–60. https://doi.org/10.2307/2405502
Barreiros JP (2016) Crocodylia: Uma longa história de sucesso evolutivo. Atlântida. Revista de Cultura 61:137–148
Bernardi G (2007) The neoselectionist theory of genome evolution. Proc Natl Acad Sci 104:8385–8390. https://doi.org/10.1073/pnas0701652104
Bista B, Valenzuela (2020) Turtle insights into the evolution of the reptilian karyotype and the genomic architecture of sex determination. Genes 11:416. https://doi.org/10.3390/genes11040416
Brochu CA (2003) Phylogenetic approaches toward Crocodylian history. Ann Rev Earth Planet Sci 31:357–397. https://doi.org/10.1146/annurev.earth.31.100901.141308
Brochu CA (2006) A new miniature horned crocodile from the Quaternary of Aldraba Atoll Western Indian Ocean. Copeia 2:149–158. https://doi.org/10.1643/0045-8511(2006)6[149:ANMHCF]2.0.CO;2
Brochu CA (2007) Morphology relationships and biogeographical significance of an extinct horned crocodile (Crocodylia Crocodylidae) from the Quaternary of Madagascar. Zool J Linn Soc 150:835–863. https://doi.org/10.1111/j.1096-3642.2007.00315.x
Bronzati M, Montefeltro FC, Langer MC (2015) Diversification events and the effects of mass extinction on Crocodyliformes evolutionary history. R Soc Open Sci 2:140385. https://doi.org/10.1098/rsos.140385
Chavananikul V, Wattanodorn S, Youngprapakorn P (1994) Karyotypes of 5 species of crocodile kept in Samutprakan Crocodile Farm and Zoo. In: Crocodiles. The 12th Working Meeting of the Crocodile Specialist Group. IUCN, Gland, pp 58–62
Cioffi MB, Liehr T, Trifonov V, Molina WF, Bertollo LAC (2013) Independent sex chromosome evolution in lower vertebrates: a molecular cytogenetic overview in the Erythrinidae fish family. Cytogenet Genome Res 141:186–194. https://doi.org/10.1159/000354039
Cohen MM, Clark HF (1967) The somatic chromosomes of five crocodilian species. Cytogenet 6:193–203. https://doi.org/10.1159/000129941
Cohen MM, Gans C (1970) The chromosomes of the order Crocodilia. Cytogenet Genome Res 9:81–105. https://doi.org/10.1159/000130080
Colston TJ, Kulkarni P, Jetz W, Pyron RA (2020) Phylogenetic and spatial distribution of evolutionary diversification, isolation, and threat in turtles and crocodilians (non-avian archosauromorphs). BMC Evol Biol 20:1–16. https://doi.org/10.1186/s12862-020-01642-3
Cronquist A (1968) The Evolution and Classification of Flowering Plants. United States of America, Boston
Deakin JE, Ezaz T (2019) Understanding the evolution of reptile chromosomes through applications of combined cytogenetics and genomic approaches. Cytogenet Genome Res 157:7–20. https://doi.org/10.1159/000495974
Degrandi TM, Barcellos SA, Costa AL, Garnero ADV, Hass I, Gunski RJ (2020) Introducing the bird chromosome database: an overview of cytogenetic studies in birds. Cytogenet Genome Res 160(4):199–205. https://doi.org/10.1159/000507768
Dobigny G, Ducroz JF, Robinson TJ, Volobouev V (2004) Cytogenetics and cladistics. Syst Biol 53:470–484. https://doi.org/10.1080/10635150490445698
Eaton MJ, Martin A, Thorbjarnarson J, Amato G (2009) Species-level diversification of African dwarf crocodiles (Genus Osteolaemus): a geographic and phylogenetic perspective. Mol Phylogenet Evol 50:496–506. https://doi.org/10.1016/j.ympev.2008.11.009
Farré M, Narayan J, Slavov GT, Damas J, Auvil L, Li C, Jarvis ED, Burt DW, Griffin DK, Larkin DM (2016) Novel insights into chromosome evolution in birds, archosaurs, and reptiles. Genome Biol Evol 8(8):2442–2251. https://doi.org/10.1093/gbe/evw166
Ferguson-Smith MA, Trifonov V (2007) Mammalian karyotype evolution. Nat Rev Genet 8:950–962. https://doi.org/10.1038/nrg2199
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specified tree topology. Syst Zool 20:406–416
Furo IO, Kretschmer R, O’Brien PCM, Pereira JC, Garnero ADV, Gunski RJ, O’Connor RE, Griffin DK, Ferguson-Smith GAJB, de Oliveira MA, EHC, (2020) Chromosomal evolution in the phylogenetic context: a remarkle haryotype reorganization in neotropical Parrot Myiopsitta monachus (Psittacidae). Front Genet 11:721. https://doi.org/10.3389/fgene.2020.00721
Gill F, Donsker D, Rasmussen P (2022) IOC World Bird List (v121). https://www.worldbirdnames.org/new/. Accessed 18 July 2022
Green RE, Braun EL, Armstrong J, Earl D, Nguyen N, Hickey G, Vandewege MW, St John JA, Capella-Gutiérrez S, Castoe TA (2014) Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346:1254449. https://doi.org/10.1126/science.1254449
Grigg G, Seebacher F, Franklin CE (2001) Crocodilian Biology and Evolution. Surrey Beatty & Sons, Chipping Norton
Grigg G, Kirshner D (2015) Biology and evolution of crocodylians. Cornell University Press, Ithaca and London
Hay JM, Sarre SD, Lambert DM, Allendorf FW, Daugherty CH (2010) Genetic diversity and taxonomy: a reassessment of species designation in tuatara (Sphenodon: Reptilia). Conserv Genet 11:1063–1081. https://doi.org/10.1007/s10592-009-9952-7
Hekkala E, Shirley MH, Amato G, Austin JD, Charter S, Thorbjarnarson J, Vliet KA, Houck ML, Desalle R, Blum MJ (2011) An ancient icon reveals new mysteries: mummy DNA resurrects a cryptic species within the Nile crocodile. Mol Ecol 20:4199–4215. https://doi.org/10.1111/j.1365-294X.2011.05245.x
Hekkala E, Gatesy J, Narechania A, Meredith R, Russelo M, Aardema ML, Jensen E, Montanari S, Brochu C, Norel M, Amato G (2021) Paleogenomics illuminates the evolutionary history of the extinct Holocene “horned” crocodile of Madagascar Voay robustus. Commun Biol 4:505. https://doi.org/10.1038/s42003-021-02017-0
Iwabe N, Hara Y, Kumazawa Y, Shibamoto K, Saito Y, Miyata T, Katoh K (2005) Sister group relationship of turtles to the bird-crocodilian clade revealed by nuclear DNA-coded proteins. Mol Biol Evol 22:810–813. https://doi.org/10.1093/molbev/msi075
Jablonski D, Roy K, Valentine JW (2006) Out of the tropics: evolutionary dynamics of the latitudinal diversity gradient. Science 314:102–106. https://doi.org/10.1126/science.1130880
Janes DE, Organ CL, Fujita MK, Shedlock AM, Edwards SV (2010) Genome evolution in Reptilia the sister group of mammals. Annu Rev Genomics Hum Genet 11:239–264. https://doi.org/10.1146/annurev-genom-082509-141646
Janke A, Arnason U (1997) The complete mitochondrial genome of Alligator mississippiensis and the separation between recent Archosauria (birds and crocodiles). Mol Biol Evol 14:1266–1272. https://doi.org/10.1093/oxfordjournals.molbev.a025736
Johnson Pokorná M, Altmanová M, Rovatsos M, Velenský P, Vodička R, Rehák I, Kratochvíl L (2016) First description of the karyotype and sex chromosomes in the Komodo dragon (Varanus komodoensis). Cytogenet Genome Res 148:284–291. https://doi.org/10.1159/000447340
Kasai F, O’Brien PCM, Martin S, Ferguson-Smith MA (2012) Extensive homology of chicken macrochromosomes in the karyotypes of Trachemys scripta elegans and Crocodylus niloticus revealed by chromosome painting despite long divergence times. Cytogenet Genome Res 136:303–307. https://doi.org/10.1159/000338111
Kawagoshi T, Nishida C, Ota H, Kumazawa Y, Endo H, Matsuda Y (2008) Molecular structures of centromeric heterochromatin and karyotypic evolution in the Siamese crocodile (Crocodylus siamensis) (Crocodylidae Crocodylia). Chromosome Res 16:1119–1132. https://doi.org/10.1007/s10577-008-1263-1
Kawai A, Nishida-Umehara C, Ishijima J, Tsuda Y, Ota H, Matsuda Y (2007) Different origins of bird and reptile sex chromosomes inferred from comparative mapping of chicken Z-linked genes. Cytogenet Genome Res 117:92–102. https://doi.org/10.1159/000103169
King M, Honeycutt R, Contreras N (1986) Chromosomal repatterning in crocodiles: C G and N-banding and the in situ hybridization of 18S and 26S rRNA cistrons. Genetica 70:191–201
Kretschmer R, Ferguson-Smith MA, de Oliveira EHC (2018) Karyotype evolution in birds: from conventional staining to chromosome painting. Genes 9:181. https://doi.org/10.3390/genes9040181
Kretschmer R, Furo IO, Cioffi MB, Gunski RJ, Garnero ADV, O’Brien PCM, Ferguson-Smith MA, de Freitas TRO, de Oliveira EHC (2020) Extensive chromosomal fissions and repetitive DNA accumulation shaped the atypical karyotypes of two Ramphastidae (Aves: Piciformes) species. Biol J Linn Soc 130:839–849. https://doi.org/10.1093/biolinnean/blaa086
Kretschmer R, Rodrigues BS, Barcellos SA, Costa AL, Cioffi MB, Garnero AV, Gunski RJ, de Oliveira EHC, Griffin DK (2021) Karyotype evolution and genomic organization of repetitive DNAs in the saffron finch Sicalis flaveola (Passeriformes Aves). Animals 11:1456. https://doi.org/10.3390/ani11051456
Lee MSY, Yates AM (2018) Tip-dating and homoplasy: reconciling the shallow molecular divergences of modern gharials with their long fossil record. Proc Royal Soc B 285:20181071. https://doi.org/10.1098/rspb.2018.1071
Lemskaya NA, Romanenko SA, Golenishchev FN, Rubtsova NV, Sablina OV, Serdukova NA, O’Brien PCM, Fu B, Yiğit N, Ferguson-Smith MA, Yang F, Graphodatsky AS (2010) Chromosomal evolution of Arvicolinae (Cricetidae Rodentia) III Karyotype relationships of ten Microtus species. Chromosome Res 18:459–471. https://doi.org/10.1007/s10577-010-9124-0
Levan A, Fredga K, Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201–220. https://doi.org/10.1111/j.1601-5223.1964.tb01953.x
Lui JF, Valencia EFT, Boer JA (1994) Karyotypic analysis and chromosome biometry of cell cultures of the yellow throated alligator (Caiman latirostris DAUDIN). Rev Bras Genet 17:165–169
Maddison WP, Maddison DR (2018) Mesquite: a modular system for evolutionary analysis. Version 3.4. https://www.mesquiteproject.org. Accessed 10 Feb 2019
Marin J, Hedges SB (2016) Time best explains global variation in species richness of amphibians birds and mammals. J Biogeogr 43:1069–1079. https://doi.org/10.1111/jbi.12709
Martin S (2008) Global diversity of crocodiles (Crocodilia Reptilia) in freshwater. Hydrobiologia 595:587–591. https://doi.org/10.1007/s10750-007-9030-4
McAliley LR, Willis RE, Ray DA, White PS, Brochu CA, Densmore LD (2006) Are crocodiles really monophyletic?—evidence for subdivisions from sequence and morphological data. Mol Phylogenet Evol 39:16–32. https://doi.org/10.1016/j.ympev.2006.01.012
McPeek MA, Brown JA (2007) Clade age and not diversification rate explains species richness among Animal taxa. Am Nat 169:4. https://doi.org/10.1086/512135
Meredith RW, Hekkala ER, Amato G, Gatesy J (2011) A phylogenetic hypothesis for Crocodylus (Crocodylia) based on mitochondrial DNA: evidence for a trans-Atlantic voyage from Africa to the New World. Mol Phylogenet Evol 60:183–191. https://doi.org/10.1016/j.ympev.2011.03.026
Nanda I, Karl E, Griffin DK, Schartl M, Schmid M (2007) Chromosome repatterning in three representative parrots (Psittaciformes) inferred from comparative chromosome painting. Cytogenet Genome Res 117:43–53. https://doi.org/10.1159/000103164
Nicolai MPJ, Matzke NJ (2019) Trait-based range expansion aided in the global radiation of Crocodylidae. Glob Ecol Biogeogr 28:1244–1258. https://doi.org/10.1111/geb.12929
Nishida C, Tsuda Y, Ishijima J, Ando J, Fujiwara A, Matsuda Y, Griffin DK (2007) The molecular basis of chromosome orthologies and sex chromosomal differentiation in palaeognathous birds. Chromosome Res 15(6):721–734. https://doi.org/10.1007/s10577-007-1157-7
Nishida C, Ishijima J, Kosaka A, Tanabe H, Habermann FA, Griffin DK, Matsuda Y (2008) Characterization of chromosome structures of Falconinae (Falconidae, Falconiformes, Aves) by chromosome painting and delineation of chromosome rearrangements during their differentiation. Chromosome Res 16:171–181. https://doi.org/10.1007/s10577-007-1210-6
Noronha RCR, Nagamachi CY, O’Brien PCM, Ferguson-Smith MA, Pieczarka JCN (2009) Neo-XY body: an analysis of XY1Y2 meiotic behavior in Carollia (Chiroptera, Phyllostomidae) by chromosome painting. Cytogenet Genome Res 124:37–43. https://doi.org/10.1159/000200086
O’Connor RE, Kiazim L, Skinner B, Fonseka G, Joseph S, Jennings R, Larkin DM, Griffin DK (2019) Patterns of microchromosome organization remain highly conserved throughout avian evolution. Chromosoma 128(1):21–29. https://doi.org/10.1007/s00412-018-0685-6
Oaks JR (2011) A time-calibrated species tree of Crocodylia reveals a recent radiation of the true crocodiles. Evolution 65:3285–3297. https://doi.org/10.1111/j.1558-5646.2011.01373.x
Oliveira VCS, Altmanová M, Viana PF, Ezaz T, Bertollo LAC, Ráb P, Liehr T, Al-Rikabi A, Feldberg E, Hatanaka T, Scholz S, Meurer A, Cioffi MB (2021) Revisiting the karyotypes of Alligators and Caimans (Crocodylia, Alligatoridae) after a half-century delay: bridging the gap in the chromosomal evolution of Reptiles. Cells 10:1397. https://doi.org/10.3390/cells10061397
Olmo E, Signorino GG (2022) Chromorep a reptile chromosomes database available online. https://www.chromorepunivpmit/?q=node/13. Accessed 24 Aug 2022
Pan T, Miao J-S, Zhang H-B, Yan P, Lee P-S, Jiang X-Y, Ouyang J-H, Dneg Y-P, Zhang B-W, Wu X-B (2021) Near-complete phylogeny of extant Crocodylia (Reptilia) using mitogenome-based data. Zool J Linn Soc 191:1075–1089. https://doi.org/10.1093/zoolinnean/zlaa074
Porter CA, Haiduk MW, de Queiroz K (1994) Evolution and phylogenetic significance of ribosomal gene location in chromosomes of squamate reptiles. Copeia 1994:302–313. https://doi.org/10.2307/1446980
Pough FH (2022) Biodiversity of Reptiles. In: Encyclopedia of Biodiversity, 3rd ed. Elsevier Inc, Dutch, pp-1–22
Rodionov AV (1996) Micro vs macro: structural-functional organization of avian micro- and macrochromosomes. Genetika 32(5):597–608
Romanenko SA, Perelman PL, Trifonov VA, Graphodatsky AS (2012) Chromosomal evolution in Rodentia. Heredity 108:4–16. https://doi.org/10.1038/hdy.2011.110. (Epub 2011 Nov 16)
Sambrook J, Russell DW (2001) Molecular Cloning: a Laboratory Manual. Cold Spring Harbor Laboratory Press, New York
Schmid M (1980) Chromosome banding in Amphibia IV differentiation of GC and AT-rich regions in Anura. Chromosoma 77:83–103
Sember A, Bertollo LAC, Ráb P, Yano CF, Hatanaka T, de Oliveira EA, Cioffi MB (2018) Sex chromosome evolution and genomic divergence in the fish Hoplias malabaricus (Characiformes Erythrinidae). Front Genet 9:71. https://doi.org/10.3389/fgene.2018.00071
Shirley MH, Vliet KA, Carr AN, Austin JD (2014) Rigorous approaches to species delimitation have significant implications for African crocodilian systematics and conservation. Proc Royal Soc B 281:20132483. https://doi.org/10.1098/rspb.2013.2483
Shirley MH, Carr AN, Nestler JH, Vliet KA, Brochu CA (2018) Systematic revision of the living African slender-snouted crocodiles (Mecistops Gray 1844). Zootaxa 4504:151–193. https://doi.org/10.11646/ZOOTAXA.4504.2.1
Simões TR, Kinney-Broderick G, Pierce SE (2022) An exceptionally preserved Sphenodon-like sphenodontian reveals deep time conservation of the tuatara skeleton and ontogeny. Commun Biol 5:195. https://doi.org/10.1038/s42003-022-03144-y
Skinner B, Griffin D (2012) Intrachromosomal rearrangements in avian genome evolution: evidence for regions prone to breakpoints. Heredity 108:37–41. https://doi.org/10.1038/hdy.2011.99
Sochorová J, Garcia S, Gálvez F, Symonová R, Kovařík A (2018) Evolutionary trends in animal ribosomal DNA loci: introduction to a new online database. Chromosoma 127:141–150. https://doi.org/10.1007/s00412-017-0651-8
Srikulnath K, Thapana W, Muangmai N (2015) Role of chromosome changes in Crocodylus evolution and diversity. Genom Inform 13:102–111. https://doi.org/10.5808/GI.2015.13.4.102
Srikulnath K, Ahmad SF, Singchat W, Panthum T (2021) Why do some vertebrates have microchromosomes? Cells 10(9):2182. https://doi.org/10.3390/cells10092182
Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75:304–306. https://doi.org/10.1016/0014-4827(72)90558-7
Uetz P, Freed P, Aguilar R, Hošek J (2023) The Reptile Database Available online. http://wwwreptile-database.org. Accessed 3 April 2023
Uno Y, Nishida C, Tarui H, Ishishita S, Takagi C, Nishimura O, Ishijima J, Ota H, Kosaka A, Matsubara K, Murakami Y, Kuratani S, Ueno N, Agata K, Matsuda Y (2012) Inference of the protokaryotypes of amniotes and tetrapods and the evolutionary processes of microchromosomes from comparative gene mapping. PLoS ONE 7:e53027. https://doi.org/10.1371/journal.pone.0053027
Valenzuela N, Adams DC (2011) Chromosome number and sex determination coevolve in turtles. Evolution 65:1808–1813. https://doi.org/10.1111/j.1558-5646.2011.01258.x
Valenzuela N, Lance VA (2004) Temperature-dependent sex determination in vertebrates. Smithsonian Books, Washington, DC
Wang J, Su W, Hu Y, Li S, O’Brien PCM, Ferguson-Smith MA, Yang F, Nie W (2022) Comparative chromosome maps between the stone curlew and three ciconiiform species (the grey heron little egret and crested ibis). BMC Ecol Evol 22:1–13. https://doi.org/10.1186/s12862-022-01979-x
Willis JC (1922) Age and area: a study of geographical distribution and origin of species. Cambridge University Press, Cambridge
Yang F, Trifonov V, Ng BL, Kosyakova N, Carter NP (2009) Generation of paint probes by flow-sorted and microdissected chromosomes. In: Liehr T (ed) Fluorescense In Situ Hybridization (FISH)—Application Guide. Springer Protocols Handbooks, Heidelberg, pp 35–32
Zwick MS, Hanson RE, Islam-Faridi MN, Stelly DM, Wing RA, Price HJ, Mcnight TD (1997) A rapid procedure for the isolation of C0t–1 DNA from plants. Genome 40:138–142. https://doi.org/10.1139/g97-020
Acknowledgements
We would like to thank the private Crocodile Zoo Protivín (www.krokodylizoo.cz), namely Miroslav Procházka, for the samples of C. mindorensis and C. moreletii, Pilsen Zoo (www.zooplzen.cz), namely Tomáš Jirásek, for samples of O. tetraspis, “maman” Colette Uwonani Ambemane (Kinshasa) for helping us to access O. osborni, and Šárka Pelikánová for her help with the laboratory work.
Funding
V.C.S.O. was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasil (CAPES) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (200401/2022–0). M.A. was supported by the Charles University Research Centre program 204069 and by the Czech Science Foundation Project No. 20-27236 J. V.G. was supported by the Czech Science Foundation (23-07331S), and the Ministry of Culture of the Czech Republic (DKRVO 2019–2023/6.VII.e, National Museum, 00023272). M.d.B.C. was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (302928/2021–9), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (2020/11772–8). MBC and TL were supported by Alexander von Humboldt-Stiftung. We acknowledge support by the German Research Foundation Projekt-Nr. 512648189 and the Open Access Publication Fund of the Thueringer Universitaets- und Landesbibliothek Jena. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasil (CAPES), Finance Code 001.
Author information
Authors and Affiliations
Contributions
V.C.S.O., M.A., V.G., R.K., and M.B.C wrote the main manuscript text; V.C.S.O., M.A., V.G., T.L., N.P., G.B., A.T., and M.B.C performed the sampling and the experiments; V.C.S.O., M.A T.E., T.L., R.U., and M.B.C. prepared the figures. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study was conducted according to the guidelines of the Ethics Committee on Animal Experimentation of the Universidade Federal de São Carlos (process number CEUA 4617090919). Collections were done under the authorization of the Chico Mendes Institute for Biodiversity Conservation (ICMBIO), System of Authorization and Information about Biodiversity (SISBIO-License No. 71857–7), and National System of Genetic Resource Management and Associated Traditional Knowledge (SISGEN-ABFF266). We further thank the CITES Authority of the Democratic Republic of the Congo for issuing permit no. CDFF2423R.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

Sup Fig. 1.
FISH with the vertebrate telomeric (TTAGGG)n probe in four representative species, namely a) Crocodylus rhombifer (2n=30), b) Crocodylus moreletii (2n=32), c) Osteolaemus osborni (2n=30) and d) Osteolaemus tetraspis (2n=38). Bar = 20 µm (PNG 519 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sales-Oliveira, V., Altmanová, M., Gvoždík, V. et al. Cross-species chromosome painting and repetitive DNA mapping illuminate the karyotype evolution in true crocodiles (Crocodylidae). Chromosoma 132, 289–303 (2023). https://doi.org/10.1007/s00412-023-00806-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-023-00806-6