Abstract
Contrasting life-history characteristics of arbuscular mycorrhizal (AM) fungal families may have important implications for mycorrhizal functioning. Nevertheless, the effect of inoculation with AM fungi having different life-history strategies on the quality parameters of tomato fruits was not investigated. In this study, fruit and sauce quality of two tomato varieties were evaluated in field conditions after inoculation with four AM fungal species belonging to Glomeraceae and Gigasporaceae. The functional relationship between AM fungal traits (i.e., root colonization structures, community diversity) and fruit quality parameters was analyzed. AM fungal inoculation increased total phenols (TPC) and lycopene concentration in fruits of both varieties (47% and 247%, respectively) and antioxidant activity in var. Rio Grande (85%). Gigasporaceae were more effective in increasing TPC and antioxidant activity compared to Glomeraceae in var. Rio Grande. Gigaspora gigantea outperformed Scutellospora pellucida in var. Pisanello for TPC, antioxidant activity, and lycopene. Inoculated strains of G. gigantea, S. pellucida, Funneliformis mosseae, and Sclerocystis sinuosa were molecularly retrieved within tomato roots. In both varieties, a functional relationship between occurrence of arbuscules in roots and fruit quality was found. In var. Rio Grande, the abundance of some native AM fungal taxa shaped the pattern of fruit quality parameters. Gigasporaceae might be of great relevance for the synthesis of health-promoting compounds in tomato and should be included in biostimulant programmes targeting the production of high-quality vegetables.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Arbuscular mycorrhizal fungi (AMF) belonging to Glomeromycota phylum (Hibbett et al. 2007; Schüβler et al. 2001; Tedersoo et al. 2018) are largely applied in horticulture as microbial biostimulants since they can lower the consumption of mineral fertilizers, as well as improve their use efficiency (Bender and van der Heijden 2015; Cavagnaro et al. 2015; Schütz et al. 2018; De Santis et al. 2022). Moreover, AMF can maintain or even enhance plant growth and food quality. In the strongly competitive fresh food market, in which greenhouse and field growers need to meet the high requests of consumers, nutritional quality and health benefits of vegetables are becoming increasingly important (Dias et al. 2012; Fanasca et al. 2006; Kyriacou and Rouphael 2018). In this context, AMF could play a pivotal role as they were reported to boost the production of health-related compounds in several vegetables, such as lettuce (Lactuca sativa L.) (Avio et al. 2017; Baslam et al. 2011a, b; 2013a, b), tomato (Solanum lycopersicum L.) (Aguilera et al. 2022; Bona et al. 2017; Carillo et al. 2020; Hart et al. 2015), pepper (Capsicum annuum L.) (Mena-Violante et al. 2006), artichoke (Cynara cardunculus L.), green asparagus (Asparagus officinalis L.) (Conversa et al. 2019), onion (Allium cepa L.) (Golubkina et al. 2020), basil (Ocimum basilicum L.), oregano (Origanum vulgare L.) (Saleh et al. 2020), and eggplant (Solanum melongena L.) (Sabatino et al. 2020). However, the output of the symbiosis may depend on factors, such as plant variety, AM fungal identity, agronomic practices, and interaction with other beneficial microorganisms and abiotic stresses (e.g. Baslam et al. 2011a,b, 2012; Baum et al. 2015; Bona et al. 2018; Carillo et al. 2020; Copetta et al. 2011; Mena-Violante et al. 2006; Saleh et al. 2020).
Tomato is one of the major vegetables in the world in term of production (FAOSTAT database, average period 2011–2020; https://www.fao.org/faostat/en/#data/QCL). It has assumed the status of “functional food” due to its high concentration in health-related biomolecules (nutraceuticals) (Dorais et al. 2008) that have been proven to reduce risk of cancer and cardiovascular diseases upon consumption (e.g., Agarwal and Rao 2000; Giovannucci 1999; Heber 2000; Toor and Savage 2005). This protective effect has been mainly attributed to the antioxidant activity performed by the carotenoid constituents of the fruits, particularly lycopene and β-carotene (Clinton 1998; Di Mascio et al. 1989; Stahl and Sies 1996). Other bioactive compounds abundant in tomato fruits are ascorbic acid and vitamin E, as well as polyphenols and terpens (Emmanuel and Babalola 2020; Gruda 2005).
The effect of AM fungal inoculation on tomato fruit quality was investigated mostly in controlled conditions, i.e., in pots in greenhouse and using single AM fungal species (i.e., Funneliformis mosseae and Rhizophagus intraradices) (Giovannetti et al. 2012; Hart et al. 2015; Nzanza et al. 2012; Ulrichs et al. 2008; Zouari et al. 2014) or mixtures of AMF from two to five species (Aguilera et al. 2022; Chouyia et al. 2022; Copetta et al. 2011; Hart et al. 2015). In such conditions, some significant increases of lycopene concentration (from 12 to 125%) (Aguilera et al. 2022; Copetta et al. 2011; Giovannetti et al. 2012; Ulrichs et al. 2008) and total phenols (9%) (Ulrichs et al. 2008) were detected in fruits. Enhanced levels of β-carotene were observed in fruits of tomato plants treated with single-inoculum (+ 45%) (Ulrichs et al. 2008) and with mixture of two AM fungal species (+ 27%) (Hart et al. 2015). Moreover, increases of total antioxidant activity in fruits of tomato inoculated with F. mosseae was only detected by Zouari et al. (2014) (+ 51%). The increases of secondary metabolites in tomato plants inoculated with F. mosseae are also supported by the results of transcriptomic analyses (Fiorilli et al. 2009; Salvioli et al. 2012; Zouari et al. 2014).
Under field conditions, where native AM fungal communities compete with inoculated AMF, the response pattern of fruit content of health-related compounds in tomato due to mycorrhizal inoculation is not clear (Bona et al. 2017, 2018; Carillo et al. 2020; Njeru et al. 2017; Pasković et al. 2021; Subramanian et al. 2006). Lycopene was variably enhanced in fruits of inoculated plants (from 5 to 47%) (Bona et al. 2017; Carillo et al. 2020), whereas was unchanged in other studies (Njeru et al. 2017; Pasković et al. 2021). Moreover, recently, increases of lipophilic and hydrophilic antioxidant activity (12% and 9%, respectively) and hydrophilic total phenols (8%) were reported in fruits of inoculated plants (Carillo et al. 2020; Pasković et al. 2021). However, these studies did not investigate the functionality of isolates beloning to different families of AMF or the changes in roots of the AM fungal communities following inoculation.
Life-history characteristics of AMF belonging to different families, such as Gigasporaceae and Glomeraceae, may have important implications for mycorrhizal functioning (e.g., Arcidiacono et al. 2023; Brundrett et al. 1999; Hart and Reader 2002, 2004; Klironomos and Hart 2002; Merryweather and Fitter 1998). Members of Gigasporaceae, extensively exploring the soil by hyphae, could give a boost to host plant growth and nutrient uptake (Chagnon et al. 2013; de Souza et al. 2005; Hart and Reader 2002; Maherali and Klironomos 2007). On the opposite side, members of Glomeraceae family, intensively colonising the root system, could have a beneficial effect on the suppression of root pathogens with a consequent strong benefit on plant growth and fruit yield. Arcidiacono et al (2023) found that tomato plants of var. Pisanello and var. Rio Grande inoculated with members of Gigasporaceae produced fruits with higher concentration of N, K, Zn, Cu, Mg, and Mn respect to those inoculated with Glomeraceae. However, there is no data available on the impact of the inoculation with AMF with different life-history strategies on the quality parameters of tomato fruits. So far, inoculation experiments were not supported by reliable molecular identification of the AM fungal taxa in roots. Indeed, in the field, the presence of the native AMF, competing with the inoculated AM fungal isolates, could have important implications on the output of the mycorrhizal symbiosis (e.g., Ercoli et al. 2017; Pellegrino et al. 2012, 2022).
The aim of this study was to investigate under field conditions how the inoculation of tomato with four single AM fungal isolates belonging to Glomeraceae and Gigasporaceae can affect the nutraceuticals content of fruits and related transformed products, such as tomato sauce. We selected two varieties of tomato, characterized by different growth habits: var. Pisanello (an old local variety) with an indeterminate growth habit and var. Rio Grande (a widely used modern variety) with a determinate growth habit. We set-up two field experiments: one in 2019 with var. Pisanello and one in 2020 with var. Rio Grande. Two AM fungal species per family were chosen to provide replication. We hypothesized the existence of functional trade-offs among AM fungal species belonging to Gigasporaceae and Glomeraceae and correlated functional traits within members of each family (Chagnon et al. 2013; Maherali and Klironomos 2007). Members of Gigasporaceae, extensively exploring the soil by hyphae, were figured to have a stronger effect on fruits nutrient uptake and consequently on the content of health-related secondary metabolites, such as lycopene, total phenols, and antioxidant activity, in comparison with members of Glomeraceae that extensively colonize the root system. We also hypothesized that development of AM fungal taxa within roots is affected by the competition between fungal species belonging to native community and inoculated species and differences at family level would be related to changes of diversity of AMF in roots, due to the expected higher colonization ability of Glomeraceae. Finally, we expected that both functional fungal traits (i.e., AM fungal abundance and community diversity) would have been highly correlated with quality parameters (i.e., lycopene, total phenols, and antioxidant activity) in tomato fruits.
Materials and methods
Characterization of the field site
The field experimental sites were located at the organic farm “Fattoria Le Prata” (Pisa, Italy) (43°44′ N, 10°24′ E; 2 m above sea level and 0.0% slope). The soil of the experiment, carried out in 2019, was a silty-clay loam (8.0% sand, 54.1% silt and 37.9% clay) 7.9 pH (deionized water 1:2.5 w/v; McLean 1982), 2.2 g kg−1 total N (Kjeldahl; Bremner and Mulvaney 1982), 1.89 g kg−1 total P, and 22.00 mg kg−1 available P (Olsen; Olsen and Sommers 1982), and with 35.9 g kg−1 soil organic C (SOC) (Walkley–Black; Nelson and Sommers 1982). The soil of the experiment, carried out in 2020, was a silty-clay loam (10.2% sand, 50.2% silt, and 39.6% clay), 8.0 pH (H2O), 1.83 g kg−1 total N (Kjeldahl), 1.72 g kg−1 total P, and 17.0 mg kg−1 available P (Olsen), with 30.5 g kg−1 soil organic C (Walkley–Black). Climate of the field area is cold, humid Mediterranean (Csa), according to the Köppen-Geiger climate classification (Kottek et al. 2006). From May to September 2019, corresponding to the field growth cycle of tomato, maximum and minimum temperatures were 27.4 °C and 16.0 °C, respectively, and total precipitation was 236 mm. From May to September 2020, maximum and minimum temperatures were 27.4 °C and 15.5 °C, respectively, while total precipitation was 270 mm. The preceding crop of tomato grown in 2019 and 2020 was bread wheat.
Fungal and plant material
The AMF used as inocula were Gigaspora gigantea PA125 and Scutellospora pellucida MN408A belonging to Gigasporaceae, and Funneliformis mosseae MD118 and Sclerocystis sinuosa MD126 belonging to Glomeraceae. Inocula were obtained from pot cultures maintained in the collection of the Crop Science Research Center of the Scuola Superiore Sant’Anna, Italy. Details about inocula are given in Table S1. The plant material used in 2019 and 2020, respectively, were Solanum lycopersicum L. var. Pisanello, an old Tuscan variety described and conserved in the regional genetic bank for the conservation of endangered (or threatened) varieties (http://germoplasma.regione.toscana.it/; Berni et al. 2018), and S. lycopersicum L. var. Rio Grande, a modern widely used variety.
Tomato inoculation with AMF at the nursery
Inoculation by AMF was performed in a climatic chamber at the sowing (var. Pisanello: 11th March 2019; var. Rio Grande: 15th April 2020), before transplanting the plantlets to the field. In detail, 160 seeds of S. lycopersicum were placed in a propagation tray with hole dimension of 26.4 mL (total volume: 4.22 L), containing as substrate a mixture (1:1:2:2 by volume) of peat, soil, coarse silica sand and heat-expanded clay. The soil used in the mixture was a sandy loam collected at the “Centro di Ricerche Agro-Ambientali Enrico Avanzi,” University of Pisa, Italy. Soil chemical and physical characteristics were 8.0 pH(deionized water 1:2.5 w/v; McLean 1982), 15.3% clay, 30.1% silt, 54.5% sand, 2.2% organic matter (Walkley–Black; Nelson and Sommers 1982), 1.3 g kg−1 total N (Kjeldahl; Bremner and Mulvaney 1982), 469.5 mg kg−1 total P (Olsen), 17.6 mg kg−1 extractable P (Olsen) (Olsen and Sommers 1982), and 149.6 mg kg−1 extractable K (Thomas 1982). The mixture was steam-sterilized (121 °C for 25 min, on two consecutive days) to kill naturally occurring AMF. The tray was inoculated either with 850 mL of crude inoculum (mycorrhizal roots and soil containing spores and extraradical mycelium) of one out of the four AMF or with 850 mL of a sterilized mixture of them (control). The non-mycorrhizal control was steam-sterilized (121 °C for 25 min, on two consecutive days). A high amount of inoculum (10% by volume) was used to balance potential differences in AM fungal colonization ability of the four inocula. Each tray received 565 mL of a filtrate (45-µm mesh size), obtained using a mixture of the four crude inocula and of a sample of the agricultural soil used in the mixture, to ensure to all treatments a common prokaryotic community. Plants were grown in a climatic chamber from the start of April to the end of May on both the years of cultivation (2019 and 2020) (24 °C and 18 °C night temperature; 14:10 h light:dark cycle, 420 µmol m−2 s−1) (ca. 60 days) and were supplied with tap water as needed and with a half-strength Hoagland solution every month (70 mL per tray).
Experimental set-up
Tomato plantlets (mean length of plantlets: 18 cm) of var. Pisanello were transplanted into the field in May (60 plants per replicate plot for var. Pisanello on16th of May 2019; 32 plants per replicate plot for var. Rio Grande on 23rd of May 2020) (a total of 900 and 480 plants, respectively). The size of the field in 2019 was 1248 m2 (52-m length × 24-m width), while the size of 2020 was 1350 m2 (90-m length × 15-m width). The infectivity of the soils before transplating was very low and similar between years (e.g., 9.1% of AM fungal root colonization assessed by the mycorrhizal infection potential test; Arcidiacono et al. 2023). A completely randomized design with five levels of the treatment AM fungal inoculation (Inoc) (G. gigantea, G.giga; S. pellucida, S.pellu; F. mosseae, F.mos; S. sinuosa, S.sin; mock-inoculated control, − M) was set up in 2019 and 2020 for var. Pisanello and Rio Grande, respectively. Schematic overview of the experimental design is given in Arcidiacono et al. (2023). Each level of Inoc was replicated three times. The replicate plot of the experiment with var. Pisanello had a size of 9.6-m length × 1.6-m width (15.4 m2) and was composed by two rows with 30 plants for each row (30 cm within the rows and 1 m between rows). Each replicate plot of the experiment with var. Rio Grande had a size of 2.5-m length × 1.25-m width (3.1 m2) and was composed by four rows with eight plants for each row (25 cm within and between the rows). In both years, the plots were separated by uninoculated plantlets of the same variety. The tomato plants were daily watered through drip irrigation, with pressure regulation, allowing similar water flow to each plot. No mineral fertilization neither chemical nor mechanical weed control were applied.
Sampling of fruits for quality assessment and of roots for molecular analysis
Harvests were performed at BBCH 89 (fully ripe: all fruits have typical fully-ripe color) (Meier 2001) on 10 plants in the central area of each plot to avoid potential border effect. Plants of var. Pisanello were harvested two times: 22nd July and 1st of August 2019. Similarly, plants of var. Rio Grande were harvested two times: 3rd August and 10th August 2020. All sampled fruits showed a ripeness stage 6 (red) (USDA 1975). Tomato fruits were washed and immediately stored at − 80 °C. Then, fruit samples were lyophilized and ground to a fine powder prior to the extraction and determination of total phenolic content (TPC), antioxidant activity, and lycopene content. In each experiment, in order to cover AM fungal spatial variability, five soil cores per each replicate plot were collected at 0–20-cm soil depth and then pooled in a combined soil sample. Sampling was carried out for both varieties at the 2nd harvest (BBCH 89; 1st of August 2019 and 10th August 2020, respectively). From each combined soil sample. roots were manually collected with forceps and washed by wet-sieving and decanting down to a mesh size of 250 mm. After removing organic debris, live and fine roots were carefully plucked with forceps and stored at − 80 °C for further analysis. Moreover, for var. Rio Grande, an additional root sampling was carried out when plantlets were transplanted in the field (BBCH 22; 23rd May 2020).
Fruit qualitative analyses
Extraction for TPC and antioxidant activity determination was done as described by Morra et al. (2021) with minor modifications. In detail, approximately 4 g of lyophilized sample (in duplicate) was homogenized in 20–25 mL of 80:20 CH3OH/H2O mixture and sonicated in an ultrasonic bath (Elmasonic P Elma, Singen, Germany) at 30 °C for 3 h into the darkness. Afterwards, the extracts were filtered and the flow through was taken and kept at − 20 °C until further analyses. The TPC was determined according to Folin–Ciocalteu method (Singleton and Rossi 1965) with minor modifications. The tube test contained 1 mL H2O, 100 μL of tomato extract and 100 μL of Folin-Ciocalteu phenol reagent 2N solution and, after 10 min, 800 μL of Na2CO3 75 g L−1 solution. The mixture was kept for 120 min at room temperature into the darkness, and the absorption was measured at 765 nm against a reagent blank. Measurements were performed in duplicate, and results were expressed as mg of gallic acid equivalent per 100 g of dry weight (mg GAE 100 g−1 d.w.). For calibration, standard solutions of gallic acid (range 0–150 μg mL−1) were prepared and estimated as above.
The antioxidant activity of fruit samples was measured using the oxygen radical absorbance capacity (ORAC) assay kit (Cell Biolabs Inc., USA). The reaction was performed according to the protocol developed by Cao et al. (1993) and validated by Cao and Prior (1999). The antioxidant standard curve was prepared using the water-soluble vitamin E analog TroloxTM. Standards (25 µL) and tenfold diluted samples (25 µL) were added into the wells of a microplate (black 96-well plates, NuncTM black microwell, Japan). Fluorescein solution, made by diluting the Fluorescein Probe 1:100 with 1X Assay Diluent, was rapidly added to each well (150 µL) using a multichannel pipette, and the plate was incubated for 30 min at 37 °C. Afterwards, Free Radical Initiator Solution (25 µL) was added into each well and the plate was immediately placed in a fluorescence microplate reader (Wallac Victor 3 Multilabel Plate Reader, PerkinElmer Inc., Waltham, MA). The fluorescence intensities were recorded every 5 min for 60–90 min at 37 °C with an excitation wavelength of 480 nm and an emission wavelength of 520 nm. Results, calculated according to the kit protocol, were expressed in µMol Trolox equivalent antioxidant capacity per g of dry weight (µMol TE g−1 d.w.).
The quantification of lycopene (ψ,ψ-carotene), was performed according to Daood et al. (2014). The samples were dissolved in acetone and n-hexane (2:1, v/v), centrifugated and filtered through a 0.45-μm pore size nylon Millipore. The extraction was performed under dark conditions to avoid lycopene isomerization and degradation. LC–MS for metabolomic analysis was performed using UHPLC-MS (Xevo TQ Absolute Triple Quadrupole Mass Spectrometry, QCA1743, Waters, Milford, MA, USA), and an electrospray ionization (ESI) probe that was operated in the positive ion mode to detect lycopene. An ACQUITY UPLC BEH C18 analytical column (2.1 mm × 100 mm, 1.7 µm particle size) was used for the analysis of the analyte present in each sample, using a column temperature of 35 °C and a flow rate of 0.8 mL/min. The solvent mixture consisted of water (solvent A) and acetone (solvent B). The gradient elution program was set as follows: 0.0–1.5 min, 80% B and 20% A; 1.5–2.5 min, 88% B and 12% A, 2.5–3.5 min, 95% B and 5% A, and finally 80% B and 20% A. The injection volume was set at 2 µL. MS parameters were as follows: the capillary voltage was set at 3.4 kV, while blocking and desolvation temperatures were set at 150 °C and 450 °C, respectively. The desolvation gas flow rate was set at 700 L/h, and the cone gas was set at 50 L/h. Cone voltages were set to 75 V, and collision energies were set to 20 eV.
Transformation of fresh tomato into tomato sauce and qualitative analyses
The tomato sauce was produced in a pilot plant at the “Fattoria Le Prata” (Pisa, Italy) from the tomato fruits collected at the 2nd harvest. Tomato samples from three replicates per each treatment were subjected to the following processing steps: (i) homogenization and blanching at 80 °C for 3 min in a stainless-steel pot; draining and passing through a sieve (hole diameter of 8 mm); filling in sterilized glass jar (volume of 500 mL); pasteurization at 92 °C for 2 min. For both varieties, the fruit qualitative analyses (TPC, antioxidant activity, and lycopene) were carried out on the tomato sauce obtained from the transformation of the fresh tomatoes.
Extraction of genomic DNA, PCR amplification, cloning, and sequencing of AM fungal community in roots of tomato
Three genomic DNA extractions from roots were performed per each replicate plot (roots were obtained from the combined soil sample obtained from each replicate plot) (Renker et al. 2006). Roots were collected at the BBCH 89 growth stage (2nd inflorescence with first flower open; Meier 2001) in both varieties as well as at BBCH 22 (2nd primary apical side shoot visible) in var. Rio Grande. Each extraction was performed from a subsample of 20 mg root dry weight using the Dneasy® Plant Mini Kit (Qiagen, Germantown, MD, USA). For var. Pisanello 45 DNA extractions were performed (three technical replicates x five levels of treatment Inoc x three plot replicates) and for var. Rio Grande 45 DNA extractions were performed at transplanting (three technical replicates x five levels of treatment Inoc x three plant replicates) and 45 DNA extractions were performed at harvest (three technical replicates x five levels of treatment Inoc x three plot replicates). From each DNA extract, a PCR amplification was performed using the primer pair the SSUmAf–LSUmAr, targeting part of the SSU, the complete ITS region (including the 5.8S rRNA gene), and approx. 0.8 kb of the LSU rRNA gene (Krüger et al. 2009). Then, SSUmCf and LSUmBr were applied as nested primers. The final concentration of the PCR reaction mix contained 0.02 U µL−1 Phusion polymerase, 1X Phusion HF Buffer with 1.5 mM MgCl2, 200 µM of each dNTP (Thermofisher, USA), and 0.5 μM of each primer, The thermal cycling was done in the S1000 Thermal Cycler™ (BIORAD, Hercules, CA, United States) in a volume of 25 µL, with the following conditions for the first PCR: initial denaturation at 98 °C for 3 min, 35 cycles at 98 °C for 10 s, primer annealing at 60 °C for 30 s, extension at 72 °C for 1 m, and a final extension at 72 °C for 10 min. The same conditions were used for the nested PCR primers except that the annealing temperature that was 63 °C and only 30 cycles. Negative and positive controls were used in each PCR cycle. The PCR products were loaded on 2% agarose gels (UltraPure Agarose, Thermofisher, USA) with 1X Tris–Borate-EDTA (TBE) at 100 V and stained with 0.5 μg mL−1 ethidium bromide. The pool of the three amplicons of DNA per each replicate was ligated into the pGem®-T Easy vector (Promega, Germany) and the production of ligation were transformed by XL10-Gold® Ultracompetent Escherichia coli cells (Agilent Technologies, Italy). On average 56 ± 2 recombinant clones per amplicon library (i.e., per replicate) were positive for the ca. 1500-bp-long fragment in var. Pisanello at 2nd harvest. For. Var. Rio Grande on average 26 ± 1 and 57 ± 4 recombinant clones per amplicon library (i.e., per replicate) were positive at transplanting and 2nd harvest, respectively. Cloning efficiency was on average 71%. Amplification was done using the GoTaq DNA Polymerase (Promega, Germany) (5 U µL−1) and primers pair SP6 and T7. PCR amplicons of the clones were visualized on agarose gels using 2% ultrapure agarose (VWR), 1X Tris–Borate-EDTA (TBE) and ethidium bromide (0.5 μg mL−1). Positive clones were converted to plasmids using the GenElute™ Plasmid Miniprep Kit (Sigma-Aldrich, USA). Sequencing reactions were set up with the vector primers SP6 and T7 using on an ABI 3730XL sequencing machine at the Eurofins Genomics (Germany).
Statistical and bioinformatic analyses
One-way analysis of variance (ANOVA) was performed on fruit quality parameters. Orthogonal contrasts were used to test differences between means: + M (all four isolates) vs. − M (mock inoculated controls) (1st comparison), Gigasporaceae vs. Glomeraceae (2nd comparison: inter-family diversity), between the two isolates of Gigasporaceae (3rd comparison: intra-family diversity), and between the two isolates of Glomeraceae (4th comparison: intra-family diversity). The effect of the transformation process (tomato fruits vs. sauce) on the fruit quality parameters of both tomato varieties was tested by one-way ANOVA, using the inoculation treatment as covariate. The data were ln- or arcsine-transformed when needed to fulfil the assumptions of ANOVA. Means and standard errors given in figures are for untransformed data. All analyses were performed using the software package on SPSS 21.0 (SPSS Inc., Chicago, IL, USA).
The Glomeromycota affiliation of the sequences was verified in similarity searches using the Basic Local Alignment Search Tool (BLASTn) in the National Center for Biotechnology Information (NCBI) database (https://blast.ncbi.nlm.nih.gov/Blast.cgi; Altschul et al. 1997). No chimeric sequences were detected among the 765, 211 and 788 newly generated AM fungal sequences (ca. 1500 bp) obtained from var. Pisanello at 2nd harvest and from var. Rio Grande at transplanting and 2nd harvest, respectively. A total number of 67 ± 1, 185 ± 7, and 61 ± 2 non-Glomeromycota sequences were obtained from var. Pisanello at 2nd harvest and from var. Rio Grande at transplanting and 2nd harvest, respectively. The AM fungal sequences obtained from roots sampled at the 2nd harvest of var. Pisanello were aligned according to Krüger et al. (2012) (http://www.amf-phylogeny.com) that allows a robust resolution and avoids artificial clustering resulting from misalignment or from convergent characters resulting from mutational saturation in the highly variable regions. The AM fungal sequences obtained from roots sampled at transplanting and at the 2nd harvest of var. Rio Grande were aligned using the same method. Three phylogenetic trees were built using the maximum likelihood (ML) analysis in MEGA11 by 100 iterations for the bootstrap method, Tamura-Nei model, and using all sites (Tamura et al. 2021). The phylograms were drawn by MEGA 11 and edited by Adobe Illustrator 2021. All representative newly generated sequences are deposited in NCBI Sequence Read (SRA) database as SUB13362899, SUB13324118, and SUB13375274.
The relative abundance of the AMF in each sample was calculated based on the ratio between the number of sequences affiliated to each phylotype and the total number of sequences obtained from the clone library. Total number of AM fungal phylotypes (Richness, S) and diversity indexes (Shannon index, H′ and Simpson index, λ) were calculated per each sample. The H′ and λ were calculated as follows: H′ = SUM(Pi*Log10(Pi)) and 1 − λ′ = 1 − SUM(Ni*(Ni − 1)/(N*(N/1)). In the formulas, Pi is the proportion of individuals belonging to the ith phylotype, Ni is the number of individuals to the ith phylotype, and N is the total number of the individuals of all phylotypes. One-way analysis of variance (ANOVA) was performed on the AM fungal phylotype richness and diversity indexes, and the differences between means were determined using orthogonal contrasts, following the pair-wise contrasts as defined above. At the 2nd harvest for Pisanello and at transplanting and 2nd harvest for Rio Grande, the cluster/similarity profile (SIMPROF) analysis was used to group the different samples into clusters based on their similarity/homogeneity of AM fungal intraradical communities (relative abundances of phylotypes) and to group the different AM fungal phylotypes based on their similarity of occurrence. Relative AM fungal abundance data were initially log(X + 1) transformed (Clarke and Warwick 2001) and the Bray–Curtis similarity was calculated. The SIMPROF cluster analysis was performed to objectively define the groups within the dendrogram. Moreover, for Pisanello and Rio Grande, the relative abundances of the AM fungal phylotypes found at 2nd harvest and at transplanting and 2nd harvest, respectively, were represented by a shade plot.
To understand in the two tomato varieties the functional relationship between AM fungal colonization traits (i.e., arbuscules, vesicles, AM fungal root colonization; data available in Arcidiacono et al. 2023) and fruit quality parameters (TPC, ORAC, and lycopene), and between AM fungal community (relative abundances of AM fungal phylotypes) and fruit quality parameters, and to understand which were the main responsible traits/phylotypes, a multivariate statistical approach was applied utilising data of samples collected at 2nd harvest. A principal component analysis (PCA) was performed on the Euclidean distance matrix of the AM fungal colonization traits that were square root-transformed data and a PCA was performed on the Euclidean distance matrix of the tomato quality parameters that were square root-transformed and standardized (Abdi and Williams 2010). A non-metric multidimensional scaling analysis (nMDS) was performed on the Bray–Curtis similarity matrix calculated on the fourth-root AM fungal relative abundances of phylotypes in inoculated and not-inoculated tomato varieties (Kruskal 1964). The relationship between AM fungal colonization traits and tomato quality parameters, and between AM fungal root community and tomato quality parameters was determined by a RELATE analysis that allows to determine the strength of the correlation between two matrices in rank-order patterns of dissimilarity (Clarke and Warwick 2001). The analysis was based on Spearman rank and 999 permutations with ρ equal to 1 representing perfect relationship. To find the best descriptor of the relationships, the BEST analysis, based on BioEnv methods (all combinations), Spearman 529 rank and 999 permutations, was applied (Clarke et al. 2008). Finally, the distance-based linear method (DistLM) analysis, using a stepwise selection and the Akaike’s information criterion (AICc), was applied to measure the significance and the variance explained by the best descriptor/s (Knorr et al. 2000), and the distance-based redundancy analysis was used to plot the first and second axes of the DistLM (Legendre and Anderson 1999).
Results
Total phenols in fruits
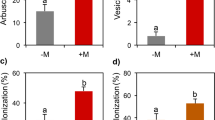
In 2019, at both 1st and 2nd harvests, AM fungal inoculation increased total phenols concentration in fruits of var. Pisanello (Fig. 1a, c; Tables S2 and S3). The increase was about 31% at 1st harvest and 45% at 2nd harvest. Similarly, in 2020, at both harvests, AM fungal inoculation increased TPC in fruits of var. Rio Grande (Fig. 1e, h), with 78 and 35% increases at 1st and 2nd harvests, respectively. A significant inter-family variability was found only in var. Rio Grande (Fig. 1f, i). At first harvest, fruits of Rio Grande inoculated with Gigasporaceae had a higher TPC than those of plants inoculated with Glomeraceae (+ 10%), whereas at 2nd harvest, the opposite behavior was observed (− 15%). In var. Pisanello, fruits of plants inoculated with G. gigantea had higher values of TPC than those of plants inoculated with S. pellucida, at both harvests, by 11 and 15%, respectively (Fig. 1b, d). By contrast, fruits of plants of var. Rio Grande inoculated with G. gigantea, at the first harvest, showed values of TPC 15% lower than those of plants inoculated with S. pellucida (Fig. 1g).
Total phenolic content (TPC) (mg of gallic acid equivalent, GAE, per 100 g of dry weight) in fruits of Solanum lycopersicum L. var. Pisanello at 1st and 2nd harvests: -M (mock inoculation, control) vs. + M (AM fungal inoculation) (n = 3 and n = 12, respectively) (a, c); Gigaspora gigantea (G.giga) vs. Scutellospora pellucida (S.pellu) (n = 3) (b, d). TPC in fruits of Solanum lycopersicum L. var. Rio Grande at 1st and 2nd harvests: − M vs. + M (n = 3 and n = 12, respectively) (e, h); Gigasporaceae (Giga) vs. Glomeraceae (Glome) (n = 6) (f, i). TPC in fruits of var. Rio Grande at first harvest: G.giga vs. S.pellu (n = 3) (g). Figure reports only the significant results among all the tested linear orthogonal contrasts (Table S2). Values are mean ± SE of three replicate plots per treatment
Antioxidant activity in fruits
In 2019, at first harvest, inoculation with G. gigantea increased by 5% the antioxidant activity (ORAC) of fruits of var. Pisanello when compared with S. pellucida (Fig. 2a, Tables S2 and S3). Accordingly, at 2nd harvest, fruits of plants of var. Rio Grande inoculated with G. gigantea showed twofold higher values of ORAC compared with those of plants inoculated with S. pellucida (Fig. 2f). In 2020, at both harvests, AM fungal inoculation increased ORAC in fruits of var. Rio Grande (Fig. 2b,d). The increase was about 122% at 1st harvest and 48% at 2nd harvest. In addition, in var. Rio Grande, a consistent inter-family variability was recorded at both harvests: higher values of ORAC where recorded in fruits of plants inoculated with Gigasporaceae compared with Glomeraceae, by 66 and 60% at 1st and 2nd harvests, respectively (Fig. 2c, e).
Antioxidant activity (oxygen radical absorbance capacity, ORAC) (µMol Trolox equivalent antioxidant capacity per g of dry weight) in fruits of Solanum lycopersicum L. var. Pisanello at first harvest: Gigaspora gigantea (G.giga) vs. Scutellospora pellucida (S.pellu) (n = 3) (a). ORAC in fruits of var. Rio Grande at 1st and 2nd harvests: − M vs. + M (n = 3 and n = 12, respectively) (b, d); Gigasporaceae (Giga) vs. Glomeraceae (Glome) (n = 6) (c, e). TPC in fruits of var. Rio Grande at 2nd harvest: G.giga vs. S.pellu (n = 3) (f). Lycopene (µg per g of dry weight) in fruits of Solanum lycopersicum L. var. Pisanello and Rio Grande at 2nd harvest: − M vs. + M (n = 3 and n = 12, respectively) (g, j); Funneliformis mosseae (F.mos) vs. Sclerocystis sinuosa (S.sin) (n = 3) (i, k). Lycopene in fruits of var. Pisanello at 2nd harvest: G.giga vs. S.pellu (n = 3) (h). Figure reports only the significant results among all the tested linear orthogonal contrasts (Table S2). Values are mean ± SE of three replicate plots per treatment
Lycopene in fruits
AM fungal inoculation greatly improved the lycopene concentration of tomato fruits of both Pisanello and Rio Grande varieties at the 2nd harvest (Fig. 2g, j; Tables S2 and S3). The increase was 344% in Pisanello and 150% in Rio Grande. Moreover, fruits of plants of var. Pisanello inoculated with G. gigantea showed higher lycopene values (+ 66%) when compared to fruits of plants inoculated with S. pellucida (Fig. 2h). Finally, a consistent intra-family variability was detected within Glomeraceae in both tomato varieties, with fruits of plants inoculated with S. sinuosa having higher values of lycopene respect to fruits of plants inoculated with F. mosseae (Fig. 2i, k). The increase was 59% in Pisanello and 118% in Rio Grande.
Tomato sauce quality parameters
AM fungal inoculation did not affect TCP, ORAC, and lycopene concentration in tomato sauce of both varieties, while differences in quality parameters at inter- and intra-family level were detected (Fig. 3; Tables S4 and S5). In var. Pisanello, lycopene was promoted in the sauce obtained from fruits of plants inoculated with Gigasporaceae in comparison with Glomeraceae (Fig. 3c; Table S4). Similarly, in var. Rio Grande, TPC and ORAC were higher in the tomato sauce from the inoculation with Gigasporaceae than in that one obtained from the inoculation with Glomeraceae (Fig. 3e,h). Conversely, for lycopene an opposite behavior was observed (Fig. 3j). In addition, an intra-family variability was found in Gigasporaceae for lycopene and TPC values of the tomato sauce of var. Pisanello and var. Rio Grande, respectively. Indeed, both quality parameters were increased by the inoculation with S. pellucida (Fig. 3d, f). Conversely, in var. Rio Grande, the ORAC was increased by the inoculation with G. gigantea (Fig. 3j). An intra-family variability was also found in Glomeraceae. Indeed, in var. Pisanello, the inoculation with S. sinuosa increased the content of TPC and ORAC in tomato sauce (Fig. 3a, b), whereas in var. Rio Grande, TPC content was increased by the inoculation with F. mosseae (Fig. 3g).
Total phenolic content (TPC) (mg of gallic acid equivalent, GAE, per 100 g of dry weight) in the tomato sauce obtained from fruits of Solanum lycopersicum L. var. Pisanello: Funneliformis mosseae (F.mos) vs. Sclerocystis sinuosa (S.sin) (n = 3) (a). TPC in the sauce of var. Rio Grande: Gigasporaceae (Giga) vs. Glomeraceae (Glome) (n = 6) (e); Gigaspora gigantea (G.giga) vs. Scutellospora pellucida (S.pellu) (n = 3) (f); F.mos vs. S.sin (n = 3) (g). Antioxidant activity (oxygen radical absorbance capacity, ORAC) (µMol Trolox equivalent antioxidant capacity per g of dry weight) in the sauce of var. Pisanello: F.mos vs S.sin (n = 3) (b). ORAC in the sauce of var. Rio Grande: Giga vs. Glome (n = 6) (h); G.giga vs. S.pellu (n = 3) (i). Lycopene (µg per g of dry weight) in the sauce of var. Pisanello: Giga vs Glome (n = 6) (c); G.giga vs. S.pellu (n = 3) (d). Lycopene (µg per g of dry weight) in the sauce of var. Rio Grande: Giga vs. Glome (n = 6) (j). Figure reports only the significant results among all the tested linear orthogonal contrasts (Table S3). Values are mean and SE of three replicate plots per treatment
Moreover, the transformation process decreased quality parameters, such as TPC and ORAC in the sauce of var. Pisanello and TPC (Fig. 4a, b; Table S6) and lycopene in the sauce of var. Rio Grande (Fig. 4d; Table S6). By contrast, lycopene in the sauce of var. Pisanello as well as ORAC in the sauce of var. Rio Grande were not significantly affected by the transformation process (Fig. 4c,e; Table S6).
Effect of the transformation process (tomato fruits vs. sauce) on the quality parameters of tomato (Solanum lycopersicum L.) var. Pisanello and var. Rio Grande: total phenolic content (TPC) (mg of gallic acid equivalent, GAE, per 100 g of dry weight) (a, d); antioxidant activity (oxygen radical absorbance capacity, ORAC) (µMol Trolox equivalent antioxidant capacity per g of dry weight) (b, e); lycopene (µg per g of dry weight) (c, f). Values are mean ± SE of 15 replicates per treatment
Diversity of AMF in roots of tomato plants
Using the small subunit-internal transcribed spacer-large subunit (SSU-ITS-LSU) rDNA sequence, foreign-inoculated strains of G. gigantea, S. pellucida, F. mosseae, and S. sinuosa were retrieved at 2nd harvest within the roots of tomato var. Pisanello (Figs. 5, S1) as well as at transplanting and 2nd harvest within the roots of tomato var. Rio Grande (Figs. 6 and 7, S2 and S3). In var. Pisanello, the phylogenetic ML tree, built aligning 765 newly generated AM fungal sequences with reference sequences of Krüger et al. (2012), allowed to discriminate a total of 11 AM fungal phylotypes (Figs. 5, S1). The AM fungal richness (S) in the inoculated roots of cv. Pisanello ranged from three to seven in G. gigantea and F. mosseae, respectively, while within the mock-inoculated roots (− M) six AM fungal phylotypes were retrieved (Table S7). Statistically significant differences were found in S, Shannon index (H′) and Simpson index (λ) between + M and − M, as well as between and within AM fungal families (Tables S7 and S8). Overall, inoculation did not determine large variation in AM fungal diversity (+ M vs. − M) (Table S7). Inoculants belonging to Glomeraceae did not modify AM fungal diversity, whereas those belonging to Gigasporaceae generally reduced diversity. Within the family Glomeraceae, F. mosseae showed a diversity pattern similar to − M. Looking at root community (Fig. S5), 45% and 29% of the AM fungal sequences retrieved in G. gigantea and S. pellucida inoculated plants were affiliated to G. gigantea and S. pellucida. Similarly, in roots inoculated with F. mosseae and S. sinuosa, 36% and 22% of the retrieved AM fungal sequences were affiliated to F. mosseae and S. sinuosa. By contrast, in mock-inoculated roots, high relative abundances of Funneliformis sp., Rhizophagus irregularis, Entrophospora claroidea, and Racocetra fulgida were retrieved, while no AM fungal sequences affiliated to G. gigantea, S. pellucida, F. mosseae, and S. sinuosa were found (Fig. S5). Consistently with the diversity patterns, the cluster analysis allowed to identify five SIMPROF supported clusters that were consistent with the AM fungal inoculation levels (Fig. 7a). However, root samples of plants inoculated with F. mosseae and S. sinuosa (Glomeraceae) showed an AM fungal community similar to the mock-inoculated control (ca. 70% and 60% similarity, respectively), whereas those inoculated with G. gigaspora and S. pellucida (Gigasporaceae) clustered together and far apart (ca. 30% similarity). The AM fungal phylotypes retrieved within the roots of var. Pisanello clustered in four different groups, irrespective to inoculation (Fig. 7a). As example, G. gigantea, R. irregularis, and S. pellucida clustered together.
Collapsed maximum likelihood (ML) tree of sequences of AMF (Glomeromycota) retrieved at 2nd harvest within the roots of tomato (Solanum lycopersicum L.) var. Pisanello inoculated with AMF at nursery and mock inoculated (control, − M). Inocula were: Gigaspora gigantea (G.giga) and Scutellospora pellucida (S.pellu) belonging to Gigasporaceae, and Funneliformis mosseae (F.mos) and Sclerocystis sinuosa (S.sin) belonging to Glomeraceae. The ML tree is based on sequences obtained from the amplification of part of the SSU, the complete ITS region, and ca. 0.8 kb of the LSU rRNA gene (ca. 1500-bp-long fragment) (Krüger et al 2009). The tree is composed by 765 newly generated sequences belonging to 11 phylotypes, plus 161 reference sequences from NCBI (see Fig. S1 for the whole ML tree). Bootstrap values (based on 1000 replicates) are shown at the nodes. The scale bar indicates substitutions per site. Portions of the ML tree are shown in (a) and (b). Collapsed phylotypes are shown by coloured branches and triangles. Pie charts represent the relative abundances of each phylotypes. The sequences of Paraglomus occultum IA702 was used as an outgroup. The newly generated sequences are highlighted in boldface, see collapsed branches
Collapsed maximum likelihood (ML) tree of sequences of AMF (Glomeromycota) retrieved at 2nd harvest within the roots of tomato (Solanum lycopersicum L.) var. Rio Grande inoculated with AMF at nursery and mock inoculated (control, − M). Inocula were Gigaspora gigantea (G.giga) and Scutellospora pellucida (S.pellu) belonging to Gigasporaceae, and Funneliformis mosseae (F.mos) and Sclerocystis sinuosa (S.sin) belonging to Glomeraceae. The ML tree is based on sequences obtained from the amplification of part of the SSU, the complete ITS region, and ca. 0.8 kb of the LSU rRNA gene (ca. 1500-bp-long fragment) (Krüger et al. 2009). The tree is composed by 788 newly generated sequences belonging to 15 phylotypes, plus 162 reference sequences from NCBI (see Fig. S4 for the whole ML tree). Bootstrap values (based on 1000 replicates) are shown at the nodes. The scale bar indicates substitutions per site. Portions of the ML tree are shown in (a), (b) and (c). Collapsed phylotypes are shown by coloured branches and triangles. Pie charts represent the relative abundances of each phylotypes. The sequences of Paraglomus occultum IA702 was used as an outgroup. The newly generated sequences are highlighted in boldface, see collapsed branches
Dendrogram of similarity profile (SIMPROF) cluster analysis (horizontal dendrogram) grouping the different samples of tomato (Solanum lycopersicum L.) var. Pisanello (a) and var. Rio Grande (b) (roots collected at 2nd harvest). Plants were inoculated with AMF at nursery and mock inoculated (control, − M). Inocula were Gigaspora gigantea (G.giga) and Scutellospora pellucida (S.pellu) belonging to Gigasporaceae, and Funneliformis mosseae (F.mos) and Sclerocystis sinuosa (S.sin) belonging to Glomeraceae. Three field replicate plots per treatment. Samples are grouped into clusters based on their similarity/homogeneity of AM fungal intraradical communities (relative abundances of phylotypes). Vertical dendrogram groups the different phylotypes based on their similarity of occurrence. Red clusters are supported by the SIMPROF analysis
At the transplanting of tomato var. Rio Grande, the phylogenetic ML tree, built aligning 211 newly generated AM fungal sequences with reference sequences of Krüger et al. (2012), allowed to successfully verify the unique presence of the AM fungal inoculants within the roots sampled in each corresponding treatment (Figs. S2 and S3). Moreover, no AM fungal sequences were retrieved in roots of mock-inoculated controls. At 2nd harvest, the phylogenetic ML tree, built aligning 788 newly generated AM fungal sequences with reference sequences of Krüger et al. (2012), allowed to discriminate a total of 15 AM fungal phylotypes (Figs. 6, S4). The AM fungal richness (S) in the inoculated roots of var. Rio Grande ranged from three to eleven in G. gigantea and F. mosseae, respectively, while within the mock-inoculated roots (− M) seven AM fungal phylotypes were retrieved (Table S7). Statistically significant differences were found in S, Shannon index (H′) and Simpson index (λ) between + M and − M, as well as between and within AM fungal families (Tables S7 and S8). Overall, inoculation did not determine large variation in AM fungal diversity (+ M vs -M) (Table S7). Inoculants belonging to Glomeraceae increased AM fungal diversity, whereas those belonging to Gigasporaceae generally reduced diversity. Within the family Glomeraceae, F. mosseae promoted the highest diversity. Looking at root community (Fig. S5), 45% and 49% of the AM fungal sequences retrieved in G. gigantea and S. pellucida inoculated plants were affiliated to G. gigantea and S. pellucida. Similarly, in roots inoculated with F. mosseae and S. sinuosa, 39% and 29% of the retrieved AM fungal sequences were affiliated to F. mosseae and S. sinuosa. By contrast, in mock-inoculated roots, high relative abundances of Funneliformis sp., Rhizophagus sp., R. irregularis, and Funneliformis coronatus were retrieved, while no AM fungal sequences affiliated to G. gigantea, S. pellucida, F. mosseae, and S. sinuosa were found (Fig. S5). Consistently with the diversity patterns, the cluster analysis allowed to identify five SIMPROF supported clusters that were consistent with the AM fungal inoculation levels (Fig. 7b). However, root samples of plants inoculated with S. sinuosa and F. mosseae (Glomeraceae) showed an AM fungal community similar to the mock-inoculated control (ca. 70%), whereas those inoculated with G. gigaspora and S. pellucida (Gigasporaceae) clustered separately and far apart (ca. 30% similarity). The AM fungal phylotypes retrieved within the roots of var. Rio Grande clustered in four different groups, irrespective to inoculation (Fig. 7b). As example, G. gigantea and R. irregularis clustered together.
Functional relationship between AM fungal traits and tomato quality parameters
Mock-inoculated controls were well separated from the inoculated ones through the PCA along the first axis, based on the AM fungal colonization parameters assessed in the roots of tomato var. Pisanello (Fig. 8a). Moreover, the PCA based on the corresponding tomato quality parameters, well separated the non-inoculated plots from the inoculated ones along the first axis (Fig. 8b). Overall, arbuscules and AM fungal colonization were positively and strongly correlated with lycopene, while vesicles with ORAC and TPC. The significance of the relationship between AM fungal colonization traits and tomato quality parameters was supported by the RELATE analysis, for which the ρ was equal to 0.30 (P = 0.028) (Fig. S6a). BEST analysis allowed to highlight that arbuscules and vesicles were good predictors (correlation:0.343; data not shown) (Table S9), while DistLM analysis supported the main role played by arbuscules in determining the pattern of quality parameters in tomato (Fig. 8e; Table S9). Similarly, in var. Rio Grande, the PCA based on the AM fungal colonization parameters, and that one based on tomato quality parameters well separated the non-inoculated plots from the inoculated ones along the first axis (Fig. 8c, d). Overall, arbuscules and AM fungal colonization were positively and strongly correlated with lycopene, while vesicles with ORAC. The significance of the relationship between AM fungal colonization traits and tomato quality parameters was supported by the RELATE analysis, for which the ρ was equal to 0.21 (P = 0.043) (Fig. S6b). BEST analysis allowed to highlight that arbuscules were good predictors (correlation:0.256; data not shown) (Table S9) and DistLM analysis supported their main role in determining the pattern of quality parameters in tomato (Fig. 8f; Table S9).
Principal component analysis (PCA) plot on AM fungal colonization traits (arbuscules, vesicles, AM fungal root colonization) in the roots of tomato (Solanum lycopersicum L.) var. Pisanello (a) and var. Rio Grande (c) at 2nd harvest. Principal component analysis (PCA) plot based on quality parameters (total phenols, TPC; antioxidant activity, ORAC; lycopene) in fruits of var. Pisanello (b) and var. Rio Grande (d) at 2nd harvest. Distance-based redundancy analysis plot used to visualize the first and second axes of the distance-based linear method (DistLM) analysis applied to measure the significance and the variance explained by the best descriptor among the AM fungal colonization traits in var. Pisanello (e) and var. Rio Grande (f)
In var. Pisanello and var. Rio Grande, nMDS plots, representing AM fungal communities within roots, showed that all treatments were well separated among each other (Fig. 9a, b). Funneliformis mosseae and S. sinuosa inoculated plants of both varieties showed root AM fungal communities similar to mock-inoculated control. Moreover, in var. Pisanello, Glomeraceae inoculated plants and control showed root AM fungal communities strongly different from those retrieved in roots of G. gigantea and S. pellucida inoculated plants (Fig. 9a). A similar pattern was observed in var. Rio Grande, although G. gigantea and S. pellucida inoculated plants had a community also distinct between each other (Fig. 9b). Looking at the RELATE analysis, no significant relationship was found between root AM fungal community and quality parameters in var. Pisanello (Rho:0.019; P = 0.376) (Fig. S6c). By contrast, in var. Rio Grande, a significant relationship was supported by the RELATE analysis, for which the ρ was equal to 0.49 (P = 0.001) (Fig. S6d). BEST analysis allowed to highlight that Archeospora sp., G. gigantea, Rhizophagus fasciculatus, and Rhizophagus sp. were good predictors (correlation:0.735; data not shown) (Table S9), and DistLM analysis supported the main role played by Archeospora sp., R. fasciculatus, and Rhizophagus sp. in determining the pattern of quality parameters in tomato (Fig. 9c; Table S9).
Non-metric multidimensional scaling analysis (nMDS) plot based on AM fungal relative abundances of phylotypes retrieved within the roots of inoculated and not-inoculated tomato (Solanum lycopersicum L.) var. Pisanello (a) and var. Rio Grande (b). Distance-based redundancy analysis plot used to visualize the first and second axes of the distance-based linear method (DistLM) analysis applied to measure the significance and the variance explained by the best descriptor among the detected AM fungal phylotypes in var. Rio Grande (c)
Discussion
Health-related compounds in fruits
In a previous work, we demonstrated the positive effect of AM fungal inoculation on yield and nutrient concentration in fruits of the tomato varieties Pisanello and Rio Grande (Arcidiacono et al. 2023). Overall, inoculants with members of Gigasporaceae were more effective in increasing the concentration of nutrients in fruits compared to Glomeraceae. However, within Gigasporaceae, the positive effect on nutrients was similar between G. gigantea and S. pellucida, whereas, within Glomeraceae, F. mosseae determined larger increases of several nutrients compared with S. sinuosa. In this work, we confirmed the positive effect of AM fungal inoculation on total phenols, antioxidant activity and lycopene in fruits. Our findings on TPC and lycopene are in accordance with previous works on several varieties of tomato (Carillo et al. 2020; Copetta et al. 2011; Giovannetti et al. 2012; Hart et al. 2015; Pasković et al. 2021; Ulrichs et al. 2008). Conversely, our results on antioxidant activity in fruits partially disagree with the results of Giovannetti et al. (2012) and Hart et al. (2015). Indeed, they reported that inoculation did not affect this parameter, whereas in our work antioxidant activity was not modified in var. Pisanello and increased in var. Rio Grande. It is also important to highlight that in the previous works the authors utilized assays based on 2,2′ azino-bis-3-ethylbenzthiazoline-6-sulfonic acid based (ABTS) and on 2,2-dipheniyl-1-picrylhydrazyl (DPHH) assays, which are less sensitive than the ORAC assay that is based on fluoresceine. It is interesting to notice that the increase in lycopene concentration showed in our work (150% and 344% in var Pisanello and var. Rio Grande, respectively) is much higher compared to the findings of Giovannetti et al. (2012) and Carillo et al. (2020), recording 19% and 47% increases, respectively. However, these authors measured lycopene on fresh basis, whereas our measures were performed on lyopilized samples.
The tested AM fungal isolates produced host benefits measured as health-related compounds variable and not fully consistent with our original hypothesis of a higher accumulation under the inoculation with members of Gigasporace. Indeed, Gigasporaceae performed better (i.e., TPC and antioxidant activity in fruits) than Glomeraceae but only in var. Rio Grande. This is the first evidence that a taxonomically-based difference has been shown looking at functions (fruit quality) other than plant growth and P uptake. Previously, Hart and Reader (2002), studying the plant response of Plantago lanceolata, Plantago major, Poa annua, and Poa pratensis inoculated with a large number of AM fungal isolates, showed that AM fungal families differed significantly in their plant benefits (host biomass). However, they found that Gigasporaceae, having the largest external mycelium, conferred less biomass promotion that Glomeraceae, having the largest internal mycelium. By contrast, similarly to our results in var. Rio Grande, they highlighted that Gigasporaceae acquired and transferred to the plant more P than Glomeraceae, but only in P. major. This supports the increased evidence of context dependency of the relation between AMF and the plant (Eom et al. 2000; van der Heijden et al. 1998). However, in addition to host specificity and intra- and extraradical mycelium traits of Gigasporaceae and Glomeraceae (Berger and Gutjahr 2021; de la Providencia et al. 2005; de Souza et al. 2005), other factors should be characterized to better explain the effect of AM fungal life-history strategy on plant response. For example, the AM fungal structures occurring within roots, such arbuscules and vesicles, and occurring in soil, such as absorptive hyphae and anastomoses, might be useful explanatory factors.
Moreover, in our study, within both AM fungal families, one isolate (i.e., G. gigantea and S. sinuosa) performed better than the other one for TPC and lycopene. This suggests that species belonging to the same family may show high functional variability. This supports previous finding on intra-family variation of plant growth and nutrient uptake (i.e., N and P) along with mycelium size and structure (Hart and Reader 2002; Munkvold et al. 2004). In addition to the intra-family functional variability, a little functional consistency was also previously found at species level (Avio et al. 2006; Börstler et al. 2008, 2010; Hart and Reader 2002; Koch et al. 2006; Mensah et al. 2015). Thus, the AM fungal variability at different taxonomic levels merits further investigation in order to better understand the complex relationship between AMF and their host plants in terms of synthesis of health-related secondary metabolites.
Health-related compounds in tomato sauce
The strong reduction observed, in this study, in TPC, antioxidant activity, and lycopene in the tomato sauce compared with fresh fruits highlights that the transformation process, based on thermal processing, draining and passing through a sieve negatively impact health-related compounds. Accordingly, Graziani et al. (2003) and Pavlović et al. (2017) reported that the antioxidant activity during thermal processing decreased compared to the value in fresh fruits of several tomato genotypes. Some studies demonstrated that lycopene is thermally stable in tomato-based food systems during mild thermal treatments (Khachik et al. 1992; Nguyen et al. 2001; Nguyen and Schwartz 1998; Thompson et al. 2000), while more intense thermal treatments can cause lycopene degradation reactions (Graziani et al 2003; Mayeaux et al. 2006; Sharma and Le Maguer 1996; Shi et al. 2003; Takeoka et al. 2001). Thus, these results support the need to develop new methods of transformation by adjusting temperature and processing time for preserving the nutraceutical quality of tomato sauce.
Overall, the increase of TPC, antioxidant activity and lycopene under inoculation with Gigasporaceae should be taken into consideration for the promotion and preservation of nutraceuticals in tomato sauce. This is the first evidence of an effect of Gigasporaceae on the concentration of secondary metabolites (nutraceuticals) in transformed products. In addition, the variability at intra-family level found in tomato fruits by our and other works (Hart and Reader 2002; Horsch et al. 2023) is here confirmed in the transformed product (sauce).
Diversity of AMF in roots of tomato plants
In this work, in accordance with Krüger et al. (2012), the phylogenetic information contained in the 3′ end of the SSU, the full ITS region and the 5′ end of the LSU (ca. 1500-bp-long fragment) was sufficient for discriminating at species level the diversity of AMF within the roots. Although this fragment does not cover the variable V4 and V5 regions of the SSU, commonly used in metabarcoding studies, and the assessment was based on a Sanger sequencing approach, the high number of screened clones per library (on average 56) together with the availability of a good number of public AM fungal sequences was sufficient to achieve robust data. The next-generation sequencing method based on Illumina platform would have allowed to sequence a higher number of reads per sample and to detect rare or low-abundance AM fungal taxa, providing a comprehensive view of the AM fungal community composition but the obtained short-reads would have allowed to assign sequences at phylogenetic resolution depending on the amplified gene region (genera or species) (Delavaux et al. 2022; Guzman et al. 2021; Higo et al. 2020; Mhlanga et al. 2022; Roy et al. 2017; Suzuki et al. 2020). By contrast, the third-generation long-read sequencing technology, such as the single molecule real-time sequencing (Pacific Biosciences, PacBio), would have enabled to obtain a lower sequencing depth compared to Illumina with an average length of 2.5 kb including the V4 and V5 regions of the SSU and providing a more efficient phylogenetic assignment (Kolaříková et al. 2021).
The molecular discrimination of the inoculated AM fungal species in the roots of both tomato varieties was certainly facilitated in our study by the fact that no AM fungal sequences affiliated to G. gigantea, S. pellucida, F. mosseae, and S. sinuosa were found in mock-inoculated controls. Indeed, in controls, the unique taxa phylogenetically close to the inoculated isolates was Funneliformis sp. close to inoculated F. mosseae. This could be determined by the fact that Gigasporaceae are adapted to live in stable and undisturbed ecosystems and are strongly reduced in tilled agricultural soils (de Souza et al. 2005; Farmer et al. 2007; Jansa et al. 2003). This depends on their peculiar life-history strategy that is characterized by the investment in somatic growth rather than in reproduction, development of large spore size and few offspring, and by a slow root infectivity rate. By contrast, members of Glomeraceae are commonly largely occurring in agricultural tilled soils due to their characteristic reproductive and colonising behavior and represented in our study up to 60% of the AM fungal community of control roots (var. Pisanello: R. irregularis 20%; Funneliformis sp. 31%; var. Rio Grande: Rhizophagus sp. 13%, R. irregularis 12%, R. fasciculatus 7%, Funneliformis sp. 18%, and F. coronatus 10%). Similarly, we found members of Glomeraceae largely occurring within the roots of inoculated treatments (on average 71% in var. Pisanello and 57% in var. Rio Grande). Funneliformis sp. was consistently present in inoculated treatments, according to its well-known generalist behavior (Moora et al. 2011; Öpik et al. 2009). The increased incidence of Glomeraceae in tilled soils has been explained by a stimulatory effect of tillage that increases the number of hyphae and colonized root fragments, acting in this AM fungal family as infective propagules, and by a lower competition for root colonization (Boddington and Dodd 2000; Jansa et al. 2003). Moreover, among Glomeraceae, S. sinuosa has been generally found at low abundance in agricultural soils (Balestrini et al. 2010; Liu et al. 2016; Turrini et al. 2017). This could explain the presence and high abundance of this AM fungal species in the plants of both tomato varieties inoculated by S. sinuosa.
The hypothesis that life-history strategies of Gigasporaceae and Glomeraceae would have differentially affected the AM fungal diversity and communities within the roots of tomato was confirmed by our results. However, members of Glomeraceae that we would expect to greatly increase the AM fungal biomass inside the roots as compared to Gigasporaceae (Hart and Reader 2002; de Souza et al. 2005) and thus strongly affect root assemblages, showed an AM fungal diversity and community like control. By contrast, members of Gigasporaceae that are expected to produce low AM fungal biomass inside the roots (Hart and Reader 2002; de Souza et al. 2005), in our study were found to reduce the AM fungal diversity and strongly modified the community in roots compared to control. These findings are supported by the data on AM fungal root colonization reported by Arcidiacono et al. (2023). Indeed, AM fungal inoculation increased root colonization irrespective of the AM fungal families in both tomato varieties. Thus, following inoculation of foreign AM fungal taxa, the competition between native AMF and inoculated taxa can be considered an important driver of the AM fungal diversity and communities in roots. A stronger competitiveness of Gigasporaceae members is evidenced by the higher relative abundance of these taxa within the roots of the varieties Pisanello and Rio Grande in comparison to Glomeraceae (up to 50% with S. pellucida in var. Rio Grande).
Functional relationship between AM fungal traits and tomato quality parameters
Following the results of Arcidiacono et al. (2023), AM fungal root colonization, arbuscules, and vesicles in var. Pisanello were significantly higher in inoculated plants than controls (88%, 360%, and 66%, respectively), and an intra-family variability in AMF root colonization of Glomeraceae was found (i.e., S. sinuosa had higher AM fungal root colonization than F. mosseae: 49%). Moreover, in var. Rio Grande, the percentage of root length containing vesicles was significantly increased by inoculation (51%), and vesicles were higher in Gigasporaceae than Glomeraceae (13%) and in F. mosseae than S. sinuosa (27%). Similarly, AM fungal root colonization was higher in plants inoculated with F. mosseae than S. sinuosa (22%). By relating the AM fungal colonization traits (i.e., arbuscules, vesicles and AM fungal root colonization) within the roots of the two tomato varieties with the corresponding fruit quality parameters (i.e., TPC, antioxidant activity, and lycopene), the change in arbuscules was sufficient to describe the pattern of fruit quality parameters. Thus, we can infer that the main determinant of the improved host benefit in Gigasporaceae-inoculated plants of var. Rio Grande is the higher occurrence of arbuscules. Indeed, inoculation by Gigasporaceae of var. Rio Grande was reported to improve, at transplanting, the occurrence of arbuscules and to increase, at harvest, nutrient concentrations in fruits compared to Glomeraceae (Arcidiacono et al. 2023). Since arbuscules are the well-known site of carbon-mineral exchanges between host and fungus (Luginbuehl et al. 2017; Parniske 2008), it is not surprising that they play this major role also in secondary metabolite synthesis by the host.
Moreover, by relating the AM fungal community in roots of the two tomato varieties with the corresponding fruit quality parameters, in var. Rio Grande, the changes of some native taxa (i.e., R. fasciculatus, Rhizophagus sp. and Archeospora sp.) were the best predictors of the pattern of quality parameters, whereas in var. Pisanello quality parameters could not be described by any AM fungal phylotypes. Thus, the main driver of the improved host benefit in var. Rio Grande is not the identity of the inoculated AM fungus, but the competition between the introduced isolate and the native AM fungal community that determines changes in the abundance of some native taxa in roots. This is in agreement with recent findings obtained by relating the AM fungal assemblages within the roots of alfalfa with corresponding plant traits (Pellegrino et al. 2022). In this work, the change in abundance of one local isolate of R. irregularis induced by all inoculation treatments was sufficient to describe the pattern of plant traits. Overall, our results suggest an environmental-driven selection for highly efficient AMF and support the use of Gigasporaceae as inoculants for enhancing the nutritional value of tomato.
Conclusions
Our work underlines the key role played by AM fungal inoculation of tomato at nursery for improving the quality parameters of fruits. The differences detected between AM fungal families in the benefit to tomato in var. Rio Grande highlight that host response, driven by life-history strategy, depends also on plant host genotype. Moreover, given the high reduced concentration of nutraceuticals in thermal-processed products, such as the tomato sauce, nursery application of selected highly efficient inoculants (i.e., Gigasporaceae) may results in a great support for the quality of the final transformed products. Nevertheless, the technology of transformation could be improved in order to optimize the beneficial outcome of the AM fungal symbiosis. Gigasporaceae were also found to be more competitive with native AMF, modifying the diversity and community in the roots of tomato, as compared to Glomeraceae. This competitive behavior, shown to be beneficial for the quality of tomato, might be risky for biodiversity losses due to bio-invasion of exotic inoculants. However, the relationship between arbuscules and fruit quality parameters in the tested tomato varieties as well as the relationship between some native AMF and the fruit quality parameters of var. Rio Grande should be verified in controlled conditions excluding confounding factors. Finally, the intra-family functional variability detected for nutraceutical compounds suggests that AM fungal family alone might not be a sufficient predictor of the host response and that further investigations are needed to better characterize the role played by intra-family variability in the outcome of symbiosis.
Data Availability
Data used in this study can be made available upon reasonable request following data-sharing regulations.
References
Abdi H, Williams LJ (2010) Principal component analysis. WIRES Comput Stat 2:433–459
Agarwal S, Rao AV (2000) Tomato lycopene and its role in human health and chronic diseases. Can Med Assoc J 163:739–744
Aguilera P, Becerra N, Alvear M, Ortiz N, Turrini A, Azcón-Aguilar C, López-Gómez M, Romero JK, Massri M, Seguel A, Mora M, Borie F (2022) Arbuscular mycorrhizal fungi from acidic soils favors production of tomatoes and lycopene concentration. J Sci Food Agric 102:2352–2358
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Arcidiacono M, Pellegrino E, Nuti M, Ercoli L (2023) Field inoculation by arbuscular mycorrhizal fungi with contrasting life-history strategies differently affects tomato nutrient uptake and residue decomposition dynamics. Plant Soil 1–23. https://doi.org/10.1007/s11104-023-05995-8
Avio L, Pellegrino E, Bonari E, Giovannetti M (2006) Functional diversity of arbuscular mycorrhizal fungal isolates in relation to extraradical mycelial networks. New Phytol 172:347–357
Avio L, Sbrana C, Giovannetti M, Frassinetti S (2017) Arbuscular mycorrhizal fungi affect total phenolics content and antioxidant activity in leaves of oak leaf lettuce varieties. Sci Hortic 224:265–271
Balestrini R, Magurno F, Walker C, Lumini E, Bianciotto V (2010) Cohorts of arbuscular mycorrhizal fungi (AMF) in Vitis vinifera, a typical Mediterranean fruit crop. Environ Microbiol Rep 2:594–604
Baslam M, Garmendia I, Goicoechea N (2011a) Arbuscular mycorrhizal fungi (AMF) improved growth and nutritional quality of greenhouse-grown lettuce. J Agric Food Chem 59:5504–5515
Baslam M, Pascual I, Sánchez-Díaz M, Erro J, García-Mina JM, Goicoechea N (2011b) Improvement of nutritional quality of greenhouse-grown lettuce by arbuscular mycorrhizal fungi is conditioned by the source of phosphorus nutrition. J Agric Food Chem 59:11129–11140
Baslam M, Erice G, Goicoechea N (2012) Impact of arbuscular mycorrhizal fungi (AMF) and atmospheric CO 2 concentration on the biomass production and partitioning in the forage legume alfalfa. Symbiosis 58:171–181
Baslam M, Esteban R, García-Plazaola JI, Goicoechea N (2013a) Effectiveness of arbuscular mycorrhizal fungi (AMF) for inducing the accumulation of major carotenoids, chlorophylls and tocopherol in green and red leaf lettuces. Appl Microbiol Biotechnol 97:3119–3128
Baslam M, Garmendia I, Goicoechea N (2013b) Enhanced accumulation of vitamins, nutraceuticals and minerals in lettuces associated with arbuscular mycorrhizal fungi (AMF): a question of interest for both vegetables and humans. Agriculture 3:188–209
Baum C, El-Tohamy W, Gruda N (2015) Increasing the productivity and product quality of vegetable crops using arbuscular mycorrhizal fungi: a review. Sci Hortic 187:131–141
Bender SF, van der Heijden MG (2015) Soil biota enhance agricultural sustainability by improving crop yield, nutrient uptake and reducing nitrogen leaching losses. J Appl Ecol 52:228–239
Berger F, Gutjahr C (2021) Factors affecting plant responsiveness to arbuscular mycorrhiza. Curr Opin Plant Biol 59:101994
Berni R, Romi M, Parrotta L, Cai G, Cantini C (2018) Ancient tomato (Solanum lycopersicum L.) varieties of tuscany have high contents of bioactive compounds. Horticulturae 4:51–64
Boddington CL, Dodd JC (2000) The effect of agricultural practices on the development of indigenous arbuscular mycorrhizal fungi. I. Field studies in an Indonesian ultisol. Plant Soil 218:137–144
Böerstler B, Thiery O, Sýkorová Z, Berner A, Redecker D (2010) Diversity of mitochondrial large subunit rDNA haplotypes of Glomus intraradices in two agricultural field experiments and two semi-natural grasslands. Mol Ecol 19:1497–1511
Bona E, Cantamessa S, Massa N, Manassero P, Marsano F, Copetta A, Lingua G, D’Agostino G, Gamalero E, Berta G (2017) Arbuscular mycorrhizal fungi and plant growth-promoting pseudomonads improve yield, quality and nutritional value of tomato: a field study. Mycorrhiza 27:1–11
Bona E, Todeschini V, Cantamessa S, Cesaro P, Copetta A, Lingua G, Gamalero E, Berta G, Massa N (2018) Combined bacterial and mycorrhizal inocula improve tomato quality at reduced fertilization. Sci Hortic 234:160–165
Börstler B, Raab PA, Thiéry O, Morton JB, Redecker D (2008) Genetic diversity of the arbuscular mycorrhizal fungus Glomus intraradices as determined by mitochondrial large subunit rRNA gene sequences is considerably higher than previously expected. New Phytol 180:452–465
Bremner JM, Mulvaney CS (1982) Nitrogen - total. In: Page AL, Miller RH, Keeney DR (eds) Methods of soil analysis, Part 2, Chemical and microbiological properties agronomy monograph, vol 9, 2nd edn. American Society of Agronomy, Madison, pp 595–624
Brundrett MC, Abbott LK, Jasper DA (1999) Glomalean mycorrhizal fungi from tropical Australia. Mycorrhiza 8:305–314
Cao G, Prior RL (1999) Measurement of oxygen radical absorbance capacity in biological samples. In: Colowick SP, Kaplan NO (eds) Methods in enzymology, vol 299. Academic Press, Cambridge, pp 50–62
Cao G, Alessio HM, Cutler RG (1993) Oxygen-radical absorbance capacity assay for antioxidants. Free Radic Biol Med 14:303–311
Carillo P, Kyratzis A, Kyriacou MC, Dell’Aversana E, Fusco GM, Corrado G, Rouphael Y (2020) Biostimulatory action of arbuscular mycorrhizal fungi enhances productivity, functional and sensory quality in ‘Piennolo del Vesuvio’Cherry tomato landraces. Agronomy 10:911
Cavagnaro TR, Bender SF, Asghari HR, van der Heijden MG (2015) The role of arbuscular mycorrhizas in reducing soil nutrient loss. Trends Plant Sci 20:283–290
Chagnon PL, Bradley RL, Maherali H, Klironomos JN (2013) A trait-based framework to understand life history of mycorrhizal fungi. Trends Plant Sci 18:484–491
Chouyia FE, Fiorentino N, Rouphael Y, Ventorino V, Fechtali T, Visconti D, Cozzolino E, Idbella M, Giordano M, Fagnano M (2022) Assessing the effect of P-solubilizing bacteria and mycorrhizal fungi on tomato yield and quality under different crop rotations. Sci Hortic 293:110740
Clarke KR, Somerfield PJ, Gorley RN (2008) Testing of null hypotheses in exploratory community analyses: similarity profiles and biota-environment linkage. J Exp Mar Biol Ecol 366:56–69
Clarke KR, Warwick R (2001) Change in marine communities: an approach 1797 to statistical analysis and interpretation. Plymouth: PRIMER-E
Clinton SK (1998) Lycopene: chemistry, biology, and implications for human health and disease. Nutr Rev 56:35–51
Conversa G, Lazzizera C, Chiaravalle AE, Miedico O, Bonasia A, La Rotonda P, Elia A (2019) Selenium fern application and arbuscular mycorrhizal fungi soil inoculation enhance Se content and antioxidant properties of green asparagus (Asparagus officinalis L.) spears. Sci Hortic 252:176–191
Copetta A, Bardi L, Bertolone E, Berta G (2011) Fruit production and quality of tomato plants (Solanum lycopersicum L.) are affected by green compost and arbuscular mycorrhizal fungi. Plant Biosyst 145:106–115
Daood HG, Bencze G, Palotas G, Pek Z, Sidikov A, Helyes L (2014) HPLC analysis of carotenoids from tomatoes using cross-linked C18 column and MS detection. J Chromatogr Sci 52:985–991
de La Providencia IE, de Souza FA, Fernández F, Delmas NS, Declerck S (2005) Arbuscular mycorrhizal fungi reveal distinct patterns of anastomosis formation and hyphal healing mechanisms between different phylogenic groups. New Phytol 165:261–271
De Santis MA, Giuliani MM, Flagella Z, Pellegrino E, Ercoli L (2022) Effect of arbuscular mycorrhizal fungal seed coating on grain protein and mineral composition of old and modern bread wheat genotypes. Agronomy 12:2418
de Souza FA, Dalpé Y, Declerck S, de la Providencia IE, Séjalon-Delmas N (2005) Life history strategies in Gigasporaceae: insight from monoxenic culture. In: Declerck S, Strullu DG, Fortin A (eds) In vitro culture of mycorrhizas. Springer-Verlag, Berlin Heidelberg, pp 73–91
Delavaux CS, Ramos RJ, Sturmer SL, Bever JD (2022) Environmental identification of arbuscular mycorrhizal fungi using the LSU rDNA gene region: an expanded database and improved pipeline. Mycorrhiza 32:145–153
Di Mascio P, Kaiser S, Sies H (1989) Lycopene as the most efficient biological carotenoid singlet oxygen quencher. Arch Biochem Biophys 274:532–538
Dias PC, Pereira MDSF, MegumiKasuya MC, Paiva HND, Oliveira LSD, Xavier A (2012) Arbuscular mycorrhizae and rhizobium in rooting and nutrition of angico-vermelhos seedlings. Rev Árvore 36:1027–1038
Dorais M, Ehret DL, Papadopoulos AP (2008) Tomato (Solanum lycopersicum) health components: from the seed to the consumer. Phytochem Rev 7:231–250
Emmanuel OC, Babalola OO (2020) Productivity and quality of horticultural crops through co-inoculation of arbuscular mycorrhizal fungi and plant growth promoting bacteria. Microbiol Res 239:126569
Eom AH, Hartnett DC, Wilson GW (2000) Host plant species effects on arbuscular mycorrhizal fungal communities in tallgrass prairie. Oecologia 122:435–444
Ercoli L, Schüßler A, Arduini I, Pellegrino E (2017) Strong increase of durum wheat iron and zinc content by field-inoculation with arbuscular mycorrhizal fungi at different soil nitrogen availabilities. Plant Soil 419:153–167
Fanasca S, Colla G, Maiani G, Venneria E, Rouphael Y, Azzini E, Saccardo F (2006) Changes in antioxidant content of tomato fruits in response to cultivar and nutrient solution composition. J Agric Food Chem 54:4319–4325
Farmer MJ, Li X, Feng G, Zhao B, Chatagnier O, Gianinazzi S, Gianinazzi-Pearson V, Van Tuinen D (2007) Molecular monitoring of field-inoculated AMF to evaluate persistence in sweet potato crops in China. Appl Soil Ecol 35:599–609
Fiorilli V, Catoni M, Miozzi L, Novero M, Accotto GP, Lanfranco L (2009) Global and cell-type gene expression profiles in tomato plants colonized by an arbuscular mycorrhizal fungus. New Phytol 184(4):975–987
Giovannetti M, Avio L, Barale R, Ceccarelli N, Cristofani R, Iezzi A, Scarpato R (2012) Nutraceutical value and safety of tomato fruits produced by mycorrhizal plants. Br J Nutr 107:242–251
Giovannucci E (1999) Tomatoes, tomato-based products, lycopene, and cancer: review of the epidemiologic literature. J Natl Cancer Inst 91:317–331
Golubkina N, Krivenkov L, Sekara A, Vasileva V, Tallarita A, Caruso G (2020) Prospects of arbuscular mycorrhizal fungi utilization in production of Allium plants. Plants 9:279
Graziani G, Pernice R, Lanzuise S, Vitaglione P, Anese M, Fogliano V (2003) Effect of peeling and heating on carotenoid content and antioxidant activity of tomato and tomato-virgin olive oil systems. Eur Food Res Technol 216:116–121
Gruda N (2005) Impact of environmental factors on product quality of greenhouse vegetables for fresh consumption. Crit Rev Plant Sci 24:227–247
Guzman A, Montes M, Hutchins L, DeLaCerda G, Yang P, Kakouridis A, Dahlquist-Willard RM, Firestone MK, Bowles T, Kremen C (2021) Crop diversity enriches arbuscular mycorrhizal fungal communities in an intensive agricultural landscape. New Phytol 231:447–459
Hart MM, Reader RJ (2002) Taxonomic basis for variation in the colonization strategy of arbuscular mycorrhizal fungi. New Phytol 153:335–344
Hart MM, Reader RJ (2004) Do arbuscular mycorrhizal fungi recover from soil disturbance differently? Trop Ecol 45:97–112
Hart MM, Ehret DL, Krumbein A, Leung C, Murch S, Turi C, Franken P (2015) Inoculation with arbuscular mycorrhizal fungi improves the nutritional value of tomatoes. Mycorrhiza 25:359–376
Heber D (2000) Colorful cancer prevention: α-carotene, lycopene, and lung cancer. Am J Clin Nutr 72:901–902
Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, Kirk TJPM, Lücking R, Lumbsch HT, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai Y-C, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Kõljalg U, Kurtzman CP, Larsson K-H, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo J-M, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio J-P, Schüssler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao Y-J, Zhang N (2007) A higher-level phylogenetic classification of the Fungi. Mycol Res 111:509–547
Higo M, Azuma M, Kamiyoshihara Y, Kanda A, Tatewaki Y, Isobe K (2020) Impact of phosphorus fertilization on tomato growth and arbuscular mycorrhizal fungal communities. Microorganisms 8:178
Horsch CC, Antunes PM, Kallenbach CM (2023) Arbuscular mycorrhizal fungal communities with contrasting life-history traits influence host nutrient acquisition. Mycorrhiza 33:1–14
Jansa J, Mozafar A, Kuhn G, Anken T, Ruh R, Sanders IR, Frossard EJEA (2003) Soil tillage affects the community structure of mycorrhizal fungi in maize roots. Ecol Appl 13:1164–1176
Khachik F, Goli MB, Beecher GR, Holden J, Lusby WR, Tenorio MD, Barrera MR (1992) Effect of food preparation on qualitative and quantitative distribution of major carotenoid constituents of tomatoes and several green vegetables. J Agric Food Chem 40:390–398
Klironomos JN, Hart MM (2002) Colonization of roots by arbuscular mycorrhizal fungi using different sources of inoculum. Mycorrhiza 12:181–184
Knorr EM, Ng RT, Tucakov V (2000) Distance-based outliers: algorithms and applications. VLDB J 8:237–253
Koch AM, Croll D, Sanders IR (2006) Genetic variability in a population of arbuscular mycorrhizal fungi causes variation in plant growth. Ecol Lett 9:103–110
Kolaříková Z, Slavikova R, Krüger C, Krüger M, Kohout P (2021) PacBio sequencing of Glomeromycota rDNA: a novel amplicon covering all widely used ribosomal barcoding regions and its applicability in taxonomy and ecology of arbuscular mycorrhizal fungi. New Phytol 231:490–499
Kottek M, Grieser J, Beck C, Rudolf B, Rubel F (2006) World map of the Köppen-Geiger climate classification updated. Meteorol Z 15:259–263
Krüger M, Stockinger H, Krüger C, Schüßler A (2009) DNA-based species level detection of Glomeromycota: one PCR primer set for all arbuscular mycorrhizal fungi. New Phytol 183:212–223
Krüger M, Krüger C, Walker C, Stockinger H, Schüßler A (2012) Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytol 193:970–984
Kruskal JB (1964) Nonmetric multidimensional scaling: a numerical method. Psychometrika 29:115–131
Kyriacou MC, Rouphael Y (2018) Towards a new definition of quality for fresh fruits and vegetables. Sci Hortic 234:463–469
Legendre P, Anderson MJ (1999) Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecological Monogr 69:1–24
Liu W, Zhang Y, Jiang S, Deng Y, Christie P, Murray PJ, Li X, Zhang J (2016) Arbuscular mycorrhizal fungi in soil and roots respond differently to phosphorus inputs in an intensively managed calcareous agricultural soil. Sci Rep 6:1–11
Luginbuehl LH, Menard GN, Kurup S, Van Erp H, Radhakrishnan GV, Breakspear A, Oldroyd GED, Eastmond PJ (2017) Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356:1175–1178
Maherali H, Klironomos JN (2007) Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316:1746–1748
Mayeaux M, Xu Z, King JM, Prinyawiwatkul W (2006) Effects of cooking conditions on the lycopene content in tomatoes. J Food Sci 71:C461–C464
McLean EO (1982) Soil pH and lime requirement. In: Page AL, Miller RH, Keeney DR (eds) Methods of soil analysis, Part 2, Chemical and microbiological properties agronomy monograph, vol 9, 2nd edn. American Society of Agronomy, Madison, MA, pp 199–224
Meier U (2001) BBCH-Monograph: growth stages of mono- and dicotyledonous plants. In: Meier U (ed) Technical Report, 2nd edn. Federal Biological Research Centre for Agriculture and Forestry, Blackwell Wiss.-Verlag, Berlin, Germany, pp 158
Mena-Violante HG, Ocampo-Jiménez O, Dendooven L, Martínez-Soto G, González-Castañeda J, Davies FT, Olalde-Portugal V (2006) Arbuscular mycorrhizal fungi enhance fruit growth and quality of chile ancho (Capsicum annuum L. cv San Luis) plants exposed to drought. Mycorrhiza 16:261–267
Mensah JA, Koch AM, Antunes PM, Kiers ET, Hart MM, Bücking H (2015) High functional diversity within species of arbuscular mycorrhizal fungi is associated with differences in phosphate and nitrogen uptake and fungal phosphate metabolism. Mycorrhiza 25:533–546
Merryweather J, Fitter A (1998) The arbuscular mycorrhizal fungi of Hyacinthoides non-scripta I. Diversity of fungal taxa. New Phytol 138:117–129
Mhlanga B, Ercoli L, Piazza G, Thierfelder C, Pellegrino E (2022) Occurrence and diversity of arbuscular mycorrhizal fungi colonising off-season and in-season weeds and their relationship with maize yield under conservation agriculture. Biol Fertil Soils 58:1–19
Moora M, Berger S, Davison J, Öpik M, Bommarco R, Bruelheide H, Kühn I, Kunin WE, Metsis M, Rortais A, Vanatoa A, Vanatoa E, Stout JC, Truusa M, Westphal C, Zobel M, Walther GR (2011) Alien plants associate with widespread generalist arbuscular mycorrhizal fungal taxa: evidence from a continental-scale study using massively parallel 454 sequencing. J Biogeogr 38:1305–1317
Morra L, Cozzolino E, Salluzzo A, Modestia F, Bilotto M, Baiano S, del Piano L (2021) Plant growth, yields and fruit quality of processing tomato (Solanum lycopersicon L.) as affected by the combination of biodegradable mulching and digestate. Agronomy 11:100
Munkvold L, Kjøller R, Vestberg M, Rosendahl S, Jakobsen I (2004) High functional diversity within species of arbuscular mycorrhizal fungi. New Phytol 164:357–364
Nelson DW, Sommers LE (1982) Total carbon, organic carbon and organic matter. In: Page AL, Miller RH, Keeney DR (eds) Methods of soil analysis, Part 2, Chemical and microbiological properties agronomy monograph, vol 9, 2nd edn. American Society of Agronomy, Madison, pp 539–579
Nguyen ML, Schwartz SJ (1998) Lycopene stability during food processing. Proc Soc Exp Biol Med 218:101–105
Nguyen M, Francis D, Schwartz S (2001) Thermal isomerisation susceptibility of carotenoids in different tomato varieties. J Sci Food Agric 81:910–917
Njeru EM, Bocci G, Avio L, Sbrana C, Turrini A, Giovannetti M, Bàrberi P (2017) Functional identity has a stronger effect than diversity on mycorrhizal symbiosis and productivity of field grown organic tomato. Eur J Agron 86:1–11
Nzanza B, Marais D, Soundy P (2012) Yield and nutrient content of tomato (Solanum lycopersicum L.) as influenced by Trichoderma harzianum and Glomus mosseae inoculation. Sci Hortic 144:55–59
Olsen SR, Sommers LE (1982) Phosphorus. In: Page AL, Miller RH, Keeney DR (eds) Methods of soil analysis, Part 2, Chemical and microbiological properties agron- omy monograph, vol 9, 2nd edn. American Society of Agronomy, Madison, pp 403–430
Öpik M, Metsis M, Daniell TJ, Zobel M, Moora M (2009) Large-scale parallel 454 sequencing reveals host ecological group specificity of arbuscular mycorrhizal fungi in a boreonemoral forest. New Phytol 184:424–437
Parniske M (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6:763–775
Pasković I, Soldo B, Ban SG, Radić T, Lukić M, Urlić B, Mimica M, Bubola KB, Colla G, Rouphael Y, Major N, Šimpraga M, Ban D, Palčić I, Franić M, Grozić K, Lukić I (2021) Fruit quality and volatile compound composition of processing tomato as affected by fertilisation practices and arbuscular mycorrhizal fungi application. Food Chem 359:129961
Pavlović N, Mladenović J, Pavlović R, Moravčević Đ, Zdravković J (2017) The impact of different thermal processing of tomato to its antioxidant activity, vitamin E, dry matter and sugar content. Food Feed Res 44:123–132
Pellegrino E, Turrini A, Gamper HA, Cafà G, Bonari E, Young JPW, Giovannetti M (2012) Establishment persistence and effectiveness of arbuscular mycorrhizal fungal inoculants in the field revealed using molecular genetic tracing and measurement of yield components. New Phytol 194:810–822
Pellegrino E, Nuti M, Ercoli L (2022) Multiple arbuscular mycorrhizal fungal consortia enhance yield and fatty acids of Medicago sativa: a two-year field study on agronomic traits and tracing of fungal persistence. Front Plant Sci 13:130
Renker C, Weißhuhn K, Kellner H, Buscot F (2006) Rationalizing molecular analysis of field-collected roots for assessing diversity of arbuscular mycorrhizal fungi: to pool, or not to pool, that is the question. Mycorrhiza 16:525–531
Roy J, Reichel R, Brüggemann N, Hempel S, Rillig MC (2017) Succession of arbuscular mycorrhizal fungi along a 52-year agricultural recultivation chronosequence. FEMS Microbiol Ecol 93:fix102
Sabatino L, Iapichino G, Consentino BB, D’Anna F, Rouphael Y (2020) Rootstock and arbuscular mycorrhiza combinatorial effects on eggplant crop performance and fruit quality under greenhouse conditions. Agronomy 10:693
Saleh AM, Abdel-Mawgoud M, Hassan AR, Habeeb TH, Yehia RS, AbdElgawad H (2020) Global metabolic changes induced by arbuscular mycorrhizal fungi in oregano plants grown under ambient and elevated levels of atmospheric CO2. Plant Physiol Biochem 151:255–263
Salvioli A, Zouari I, Chalot M, Bonfante P (2012) The arbuscular mycorrhizal status has an impact on the transcriptome profile and amino acid composition of tomato fruit. BMC Plant Biol 12:1–12
Schütz L, Gattinger A, Meier M, Müller A, Boller T, Mäder P, Mathimaran N (2018) Improving crop yield and nutrient use efficiency via biofertilization-A global meta-analysis. Front Plant Sci 8:2204
Schüβler A, Schwarzott D, Walker C (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105:1413–1421
Sharma SK, Le Maguer M (1996) Kinetics of lycopene degradation in tomato pulp solids under different processing and storage conditions. Food Res Int 29:309–315
Shi J, Le Maguer M, Bryan M, Kakuda Y (2003) Kinetics of lycopene degradation in tomato puree by heat and light irradiation. J Food Process Eng 25:485–498
Singleton VL, Rossi JA (1965) Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am J Enol Vitic 16:144–158
Stahl W, Sies H (1996) Lycopene: a biologically important carotenoid for humans? Arch Biochem Biophys 336:1–9
Subramanian KS, Santhanakrishnan P, Balasubramanian P (2006) Responses of field grown tomato plants to arbuscular mycorrhizal fungal colonization under varying intensities of drought stress. Sci Hortic 107:245–253
Suzuki K, Takahashi K, Harada N (2020) Evaluation of primer pairs for studying arbuscular mycorrhizal fungal community compositions using a MiSeq platform. Biol Fertil Soils 56:853–858
Takeoka GR, Dao L, Flessa S, Gillespie DM, Jewell WT, Huebner B, Bertow D, Ebeler SE (2001) Processing effects on lycopene content and antioxidant activity of tomatoes. J Agric Food Chem 49:3713–3717
Tamura K, Stecher G, Kumar S (2021) MEGA 11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027
Tedersoo L, Sánchez-Ramírez S, Kõljalg U, Bahram M, Döring M, Schigel D, May T, Ryberg M, Abarenkov K (2018) High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers 90:135–159
Thomas GW (1982) Exchangeable cations. In: Page AL, Miller RH, Keeney DR (eds) Methods of soil analysis, Part 2, Chemical and microbiological properties agronomy monograph, vol 9, 2nd edn. American Society of Agronomy, Madison, pp 159–165
Thompson KA, Marshall MR, Sims CA, Wei CI, Sargent SA, Scott JW (2000) Cultivar, maturity, and heat treatment on lycopene content in tomatoes. J Food Sci 65:791–795
Toor RK, Savage GP (2005) Antioxidant activity in different fractions of tomatoes. Food Res Int 38:487–494
Turrini A, Agnolucci M, Palla M, Tomé E, Tagliavini M, Scandellari F, Giovannetti M (2017) Species diversity and community composition of native arbuscular mycorrhizal fungi in apple roots are affected by site and orchard management. Appl Soil Ecol 116:42–54
Ulrichs C, Fischer G, Büttner C, Mewis I (2008) Comparison of lycopene, β-carotene and phenolic contents of tomato using conventional and ecological horticultural practices, and arbuscular mycorrhizal fungi (AMF). Agron Colomb 26:40–46
USDA (1975) United States Standards for Grades of Fresh Tomatoes. Available online at: https://www.ams.usda.gov/sites/default/files/media/Tomato_Standard%5B1%5D.pdf. Accessed 1 July 2019
van der Heijden MG, Boller T, Wiemken A, Sanders IR (1998) Different arbuscular mycorrhizal fungal species are potential determinants of plant community structure. Ecology 79:2082–2091
Zouari I, Salvioli A, Chialva M, Novero M, Miozzi L, Tenore GC, Bagnaresi P, Bonfante P (2014) From root to fruit: RNA-Seq analysis shows that arbuscular mycorrhizal symbiosis may affect tomato fruit metabolism. BMC Genom 15:1–19
Acknowledgements
The authors would like to thank Alessandro Agostini Venerosi della Seta for allowing to run the field experiments at the “Fattoria Le Prata” farm (Pisa, Italy).
Funding
Open access funding provided by Scuola Superiore Sant'Anna within the CRUI-CARE Agreement. This work was funded by the the European Agricultural Fund for Rural Development 2007–2013 for Tuscany (Italy), measure 16.1 (FERTIBIO project; CUP Artea 743548) project leader Dr. Elisa Pellegrino and measure 16.2 (FERTIBIO project; CUP Artea 828090) project leader Prof. Laura Ercoli. Myriam Arcidiacono was supported by a PhD scholarship from Scuola Superiore Sant’Anna (SSSA, Pisa, Italy).
Author information
Authors and Affiliations
Contributions
EP and LE acquired funding, conceived, and designed the study. MA carried out the experimental procedures. EP and MA performed the molecular study, collected the data, performed the statistical analyses, and wrote the first draft of the manuscript. AF set-up the extractions and following analysis of antioxidant activities. LE and AF contributed to the discussion of the data. All authors commented on the manuscript and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pellegrino, E., Arcidiacono, M., Francini, A. et al. Arbuscular mycorrhizal fungi with contrasting life-history strategies differently affect health-promoting compounds in field-grown tomato by changing arbuscule occurrence and mycorrhizal assemblages in roots. Biol Fertil Soils 60, 115–136 (2024). https://doi.org/10.1007/s00374-023-01778-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-023-01778-6