Abstract
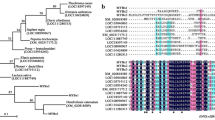
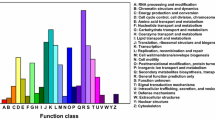
Three ANTHOCYANIN SYNTHESIS REGULATOR genes (ASR1–3) that encode R2R3-MYB transcription factors were identified recently from Petunia hybrida. In this study, we conducted additional experiments to characterize their specific function in the regulation of anthocyanin synthesis. The ASR1–3 proteins were localized in the nucleus. Analysis of their regulatory function by transient expression in the petunia corolla and yeast two-hybrid assays showed that the residues Arg at position 51 and Ala at position 102 in the N-terminal domain were essential for the regulatory function of the proteins, and the conserved domain at the C-terminal end was important for activation of the protein. RNA sequencing of overexpression (OE) and RNA-interference transgenic lines confirmed that the ASR proteins specifically induce structural genes of anthocyanin synthesis, including the early biosynthesis genes CHSj, F3H, and F3′5′H-1, the late biosynthesis genes DFR, ANS, RT, MT, AT, and GT, and the anthocyanin-related glutathione S-transferase gene AN9. In addition, a member of the detoxifying efflux carrier family, a DTX35-like gene, was upregulated by ASRs. Determination of anthocyanin/anthocyanidin contents in ASR1-OE petunia lines revealed that ASR1 especially induced the flavonoid 3′,5′-hydroxylase, which was consistent with the abundance of dihydromyricetins and delphinidins, but not cyanidins. The ASR genes were induced under high-intensity light and upregulated the expression of MYBx and MYB27, thereby providing feedback repression of anthocyanin synthesis. An updated model is presented outlining the mechanisms underlying anthocyanin synthesis in petunia.







Similar content being viewed by others
Abbreviations
- AN:
-
Anthocyanin
- ANS:
-
Anthocyanidin synthase
- AT:
-
Acylation
- bHLH:
-
Basic helix-loop-helix
- 4CL:
-
4-Coumaroyl-CoA ligase
- C4H:
-
Cinnamate 4-hydroxylase
- CHI:
-
Chalcone isomerase
- CHS:
-
Chalcone synthase
- DFR:
-
Dihydroflavonol 4-reductase
- DHK:
-
Dihydrokaempferol
- DHM:
-
Dihydromyricetin
- DHQ:
-
Dihydroquercetin
- DPL:
-
DEEP PURPLE
- F3H:
-
Flavanone 3-hydroxylase
- F3′H:
-
Flavonoid 3′-hydroxylase
- F3′5′H:
-
Flavonoid 3′,5′-hydroxylase
- FLS:
-
Flavonol synthase
- GT:
-
Glucosylation
- MT:
-
Methylation
- PAC:
-
Pale Aleurone Color
- PAL:
-
Phenylalanine ammonia lyase
- PAP:
-
PRODUCTION OF ANTHOCYANIN PIGMENT
- PHZ:
-
PURPLE HAZE
- R2R3-MYB:
-
R2R3 repeat myeloblastosis protein
- RT:
-
Rhamnosylation
- SD:
-
Synthetic dropout medium
- TT:
-
Transparent testa
- TTG:
-
Transparent testa glabra
- WDR:
-
Beta-transducin repeat protein
References
Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE (2014a) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26(3):962–980
Albert NW, Davies KM, Schwinn KE (2014b) Gene regulation networks generate diverse pigmentation patterns in plants. Plant Signaling and Behavior 9(9):e29526
Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM (2011) Members of an R2R3-MYB transcription factor family in petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J 65(5):771–784
Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13(3):99–102
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol. 11:106–106
Bliek M, Spelt K, Passeri V, Urbanus SL, Koes R, Quattrocchio FM. (2016) The genes behind the different colors of P. axillaris and P. inflata flowers. Nature Plants (Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrid, Supplemental Note 7)
Buer CS, Imin N, Djordjevic MA (2010) Flavonoids: new roles for old molecules. J Integr Plant Biol 52(1):98–111
Bombarely A, Moser M, Amrad A, Bao M, Bapaume L et al (2016) Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nature Plants 2(6):16074
Chen S, Matsubara K, Omori T, Kokubun H, Kodama H, Watanabe H, Hashimoto G, Marchesi E, Bullrich L, Ando T (2007) Phylogenetic analysis of the genus Petunia (Solanaceae) based on the sequence of the Hf1gene. J Plant Res 120(3):385–397
Consonni G, Geuna F, Gavazzi G, Tonelli C (1993) Molecular homology among members of the R gene family in maize. Plant J 3(2):335–346
Conner AJ, Albert NW, Deroles SC (2009) Transformation and regeneration of petunia. In: Gerats T, Strommer J (eds) Petunia: Evolutionary, developmental and physiological genetics. Springer, New York, pp 395–409
de Vetten N, Quattrocchio F, Mol J, Koes R (1997) The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev 1(11):1422–1434
Fu Z, Wang L, Shang H, Dong X, Jiang H, Zhang J, Wang H, Li Y, Yuan X, Meng S, Gao J, Feng N, Zhang H (2019) An R3-MYB gene of Phalaenopsis, MYBx1, represses anthocyanin accumulation. Plant Growth Regul 88(2):129–138
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827
Gould KS (2004) Nature’s Swiss army knife: the diverse protective roles of anthocyanins in leaves. J Biomed Biotechenol 5:314–320
Hoballah ME, Gubitz T, Stuurman J, Broger L, Barone M, Mandel T, Dell'Olivo A, Arnold M, Kuhlemeier C (2007) Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell 19:779–790
Holton TA, Brugliera F, Lester DR, Tanaka Y, Hyland CD, Menting JG, Lu CY, Farcy E, Stevenson TW, Cornish EC (1993) Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 366(6452):276–279
Hsu CC, Chen YY, Tsai WC, Chen WH, Chen HH (2015) Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol 168(1):175–191
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M (2019) New approach for understanding genome variations in KEGG. Nucleic Acids Res 47:D590–D595
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10(5):236–242
Kroon AR (2004) Transcription regulation of the anthocyanin pathway in Petunia hybrida. Vrije Universiteit, Amsterdam
Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57:405–430
Maier A, Schrader A, Kokkelink L, Falke C, Welter B, Iniesto E, Rubio V, Uhrig JF, Hülskamp M, Hoecker U (2013) Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J 74:638–651
Martins TR, Berg JJ, Blinka S, Rausher MD, Baum DA (2013) Precise spatiotemporal regulation of the anthocyanin biosynthetic pathway leads to petal spot formation in Clarkia gracilis (Onagraceae). New Phytol 197:958–969
Matsubara K, Kodama H, Kokubun H, Watanabe H, Ando T (2005) Two novel transposable elements in a cytochrome P450 gene govern anthocyanin biosynthesis of commercial petunias. Gene 358:121–126
Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R (1999) Molecular analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color. Plant Cell 11:1433–1444
Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10(2):63–70
Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR (2013) The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol Bioch 72:21–34
Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18(4):831–851
Spelt C, Quattrocchio F, Mol JN, Koes R (2000) Anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structuralanthocyanin genes. Plant Cell 12(9):1619–1632
Steyn WJ, Wand SJE, Holcroft DM, Jacobs G (2002) Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Physiologist 155:349–361
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4(5):447–456
Takos AM, Jaffe FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142:1216–1232
Tanaka Y, Brugliera F (2013) Flower colour and cytochromes P450. Philos Trans R Soc Lond B Biol Sci 368(1612):2012043
Tanaka T, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54:733–749
Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139:1840–1852
Tornielli G, Koes R, Quattrocchio F (2008) The genetics of flower color. In: Strommer J (ed) Petunia: Evolutionary, Developmental and Physiological Genetics (Gerats, T. Springer, New York, NY, pp 269–299
Verweij W, Spelt C, Di Sansebastiano GP, Vermeer J, Reale L, Ferranti F, Koes R, Quattrocchio F (2008) An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nat Cell Biol 10(12):1456–1462
Wang N, Xu H, Jiang S, Zhang Z, Lu N, Qiu H, Qu C, Wang Y, Wu S, Chen X (2017) MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f niedzwetzkyana). Plant J 90(2):276–292
Walker AR, Lee E, Bogs J, McDavid DA, Thomas MR, Robinson SP (2007) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J 49(5):772–785
Winkel-Shirley B (2001) Flavonoid biosynthesis: A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126(2):485–493
Willmer P (2011) Pollination and floral ecology. Princeton University Press, Princeton, New Jersey
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20(3):176–185
Yamagishi M, Shimoyamada Y, Nakatsuka T, Masuda K (2010) Two R2R3-MYB genes, homologs of Petunia AN2, regulate anthocyanin biosyntheses in flower Tepals, tepal spots and leaves of Asiatic hybrid lily. Plant Cell Physiol 51:463–474
Zhang H, Koes R, Shang H, Fu Z, Wang L, Dong X, Zhang J, Passeri V, Li Y, Jiang H, Gao J, Li Y, Wang H, Quattrocchio FM (2019) Identification and functional analysis of three new anthocyanin R2R3-MYB genes in Petunia. Plant Direct 3(1):e00114
Zhu HF, Fitzsimmons K, Khandelwal A, Kranz RG (2009) CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Molecular Plant 2:790–802
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40:22–34
Acknowledgements
We thank Dr Ronald Koes and Francesca M. Quattrocchio for the petunia cultivars and Gateway vectors. We thank Robert McKenzie, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (U1504320) and Financial Project of Henan Province (20188105 and 2019XKYH03).
Author information
Authors and Affiliations
Contributions
HZ designed the experiments and wrote the manuscript; ZF, HJ, and LW performed the gene searches and RNA-Seq data analysis; XD, and HW cared for the plants; JZ and JG performed DNA and RNA extraction; YC, YL, and MX performed qRT-PCR analysis and generated transgenic petunia plants.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fu, Z., Jiang, H., Chao, Y. et al. Three Paralogous R2R3-MYB Genes Contribute to Delphinidin-Related Anthocyanins Synthesis in Petunia hybrida. J Plant Growth Regul 40, 1687–1700 (2021). https://doi.org/10.1007/s00344-020-10224-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-020-10224-y