Abstract
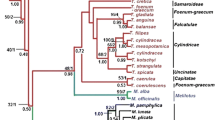
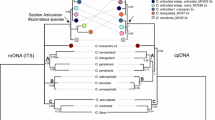
Polymerase chain reaction fragment length polymorphisms and nucleotide sequences for a cytochrome P450 gene encoding flavonoid-3′,5′-hydroxylase, Hf1, were studied in 19 natural taxa of Petunia. Natural Petunia taxa were classified into six groups based on major insertion or deletion events that occurred only in intron II of the locus. The maximum parsimony method was used to calculate strict consensus trees based on nucleotide sequences in selected regions of the Hf1 locus. Petunia taxa were divided into two major clades in the phylogenetic trees. Petunia axillaris (including three subspecies), P. exserta, and P. occidentalis formed a clade with 100% bootstrap support. This clade is associated with a consistently inflexed pedicel, self-compatibility in most taxa, and geographical distribution in southern and western portions of the genus range. The other clade, which comprised the remainder of the genus is, however, less supported (up to 71% bootstrap); it is characterized by a deflexed pedicel in the fruiting state (except P. inflata), self-incompatibility, and a northeastern distribution. A nuclear gene, Hf1, seems to be a useful molecular marker for elucidating the phylogeny of the genus Petunia when compared with the nucleotide sequence of trnK intron of chloroplast DNA.






Similar content being viewed by others
References
Ando T (1996) Distribution of Petunia axillaris (Solanaceae) and its new subspecies in Argentina and Bolivia. Acta Phytotaxon Geobot 47:19–30
Ando T, Hashimoto G (1993) Two new species of Petunia (Solanaceae) from southern Brazil. Bot J Linn Soc 111:265–280
Ando T, Hashimoto G (1994) A new Brazilian species of Petunia (Solanaceae) from the Serra da Mantiqueira. Brittonia 46:340–343
Ando T, Hashimoto G (1995) Petunia guarapuavensis (Solanaceae): a new species from Planalto of Paraná and Santa Catarina, Brazil. Brittonia 47:328–334
Ando T, Hashimoto G (1996) A new Brazilian species of Petunia (Solanaceae) from interior Santa Catarina and Rio Grande do Sul, Brazil. Brittonia 48:217–223
Ando T, Hashimoto G (1998) Two new species of Petunia (Solanaceae) from southern Rio Grande do Sul, Brazil. Brittonia 50:483–492
Ando T, Kurata M, Sasaki S, Ueda Y, Hashimoto G, Marchesi E (1995) Comparative morphological studies on infraspecific taxa of Petunia integrifolia (Hook.) Schinz et Thell. (Solanaceae). J Jpn Bot 70:205–217
Ando T, Tsukamoto T, Akiba N, Kokubun H, Watanabe H, Ueda Y, Marchesi E (1998) Differentiation in the degree of self-incompatibility in Petunia axillaris (Solanaceae) occurring in Uruguay. Acta Phytotaxon Geobot 49:37–47
Ando T, Saito N, Tatsuzawa F, Kakefuda T, Yamakage K, Ohtani E, Koshiishi M, Matsusake Y, Kokubun H, Watanabe H, Tsukamoto T, Ueda Y, Hashimoto G, Marchesi E, Asakura K, Hara R, Seki H (1999) Floral anthocyanins in wild taxa of Petunia (Solanaceae). Biochem Syst Ecol 27:623–650
Ando T, Kokubun H, Watanabe H, Tanaka N, Yukawa T, Hashimoto G, Marchesi E, Suárez E, Basulado I (2005a) Phylogenetic analysis of Petunia sensu Jussieu (Solanaceae) using chloroplast DNA RFLP. Ann Bot 96:289–297. doi:10.1093/aob/mci177
Ando T, Ishikawa N, Watanabe H, Kokubun H, Yanagisawa Y, Hashimoto G, Marchesi E, Suárez E (2005b) A morphological study of the Petunia integrifolia complex (Solanaceae). Ann Bot 96:887–900. doi:10.1093/aob/mci241
Ando T, Soto S, Suárez E (2005c) New records of Petunia (Solanaceae) for the Argentinean flora. Darwiniana 43:64–68
Cerny TA, Caetano-Anollés G, Trigiano RN, Starman TW (1996) Molecular phylogeny and DNA amplification fingerprinting of Petunia taxa. Theor Appl Genet 92:1009–1016. doi:10.1007/BF00224042
Cornu A, Maizonnier D (1983) The genetics of petunia. In: Janick J (ed) Plant breeding reviews, vol 1. AVI Publishing Company, Westport, pp 11–58
Fries RE (1911) Die Arten der Gattung Petunia. Kungl Svenska Vetensk Akad Handl 46:1–72
Griesbach RJ, Beck RM, Stehmann JR (2000) Molecular heterogeneity of the chalcone synthase intron in Petunia. HortScience 35:1347–1349
Holmgren PK, Holmgren NH, Barnett LC (1990) Index herbariorum Part I: the herbaria of the world. New York Botanical Garden, Bronx
Holton TA, Brugliera F, Lester DR, Tanaka Y, Hyland CD, Menting JGT, Lu C-Y, Farcy E, Stevenson TW, Cornish EC (1993) Cloning and expression of cytochrome P450 genes controlling flower color. Nature 366:276–279
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071–1083
Johnson LA, Soltis DE (1994) matK DNA sequences and phylogenetic reconstruction in Saxifragaceae s. str. Syst Bot 19:143–156
Jussieu AL (1803) Sur le Petunia, genre nouveau de la famille des plantes solanées. Ann Mus Natl d’Histoire Naturelle 2:1347–1349
Kabbaj A, Zeboudj F, Peltier D, Tagmount A, Tersac M, Delieu H, Berbillé A (1995) Variation and phylogeny of the ribosomal DNA unit types and 5S DNA in Petunia Jussieu. Genet Resour Crop Evol 42:311–325
Kidwell M, Lisch D (1997) Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci USA 94:7704–7711
Kokubun H, Nakano M, Tsukamoto T, Watanabe H, Hashimoto G, Marchesi E, Bullrich L, Basualdo IL, Kao T-h, Ando T (2006) Distribution of self-compatible and self-incompatible populations of Petunia axillaris (Solanaceae) outside Uruguay. J Plant Res 119:419–430. doi:10.1007/s10265-006-0002-y
Kulcheski FR, Muschner VC, Lorenz-Lemke AP, Stehmann JR, Bonatto SL, Salzano FM, Freitas LB (2006) Molecular phylogenetic analysis of Petunia Juss. (Solanaceae). Genetica 126:3–14. doi:10.1007/s10709-005-1427-2
Lassner MW, Peterson P, Yoder JI (1989) Simultaneous amplification of multiple DNA fragments by polymerase chain reaction in the analysis of transgenic plants and their progeny. Plant Mol Biol Rep 7:116–128
Matsubara K, Kodama H, Kokubun H, Watanabe H, Ando T (2005) Two novel transposable elements in a cytochrome P450 gene govern anthocyanin biosynthesis of commercial petunias. Gene 358:121–126. doi:10.1016/j.gene.2005.05.031
Quattrocchio F, Wing J, Woude KVD, Souer E, Vettern ND, Mol J, Koes R (1999) Molecular analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color. Plant Cell 11:1433–1444
Rønsted N, Law S, Thornton H, Fay MF, Chase MW (2005) Molecular phylogenetic evidence for the monophyly of Fritillaria and Lilium (Liliaceae; Liliales) and the infrageneric classification of Fritillaria. Mol Phylogenet Evol 35:509–527
Rowold DJ, Herrera RJ (2000) Alu elements and the human genome. Genetica 108:57–72
Sang T (2002) Utility of low-copy nuclear gene sequences in plant phylogenetics. Crit Rev Biochem Mol Biol 37:121–147
Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W-S, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 92:142–166
Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Syst Biol 49:369–381
Smith LB, Downs RJ (1964) Notes on the Solanaceae of southern Brazil. Phytologia 10:422–453
Smith LB, Downs RJ (1966) Petunia. In: Reitz PR (ed) Flora Illustrada Catarinense. Solanaceas. Herbário “Barbosa Rodrigues”, Itajai, Santa Catarina, Itajai, Brazil, pp 261–291
Stehmann JR (1987) Petunia exserta (Solanaceae): Uma nova espécie do Rio Grande do Sul, Brasil. Napaea Revista Bot 2:19–21
Swofford DL (1998) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Tsukamoto T, Ando T, Kokubun H, Watanabe H, Tanaka R, Hashimoto G, Marchesi E, Kao T.-h (1998a) Differentiation in the status of self-incompatibility among all natural taxa of Petunia (Solanaceae). Acta Phytotaxon Geobot 49:115–133
Tsukamoto T, Ando T, Kurata M, Watanabe H, Kokubun H, Hashimoto G, Marchesi E (1998b) Resurrection of Petunia occidentalis R. E. Fr. (Solanaceae) inferred from a cross-compatibility study. J Jpn Bot 73:15–21
Ugarkovic D, Plohl M (2002) Variation in satellite DNA profiles—causes and effects. EMBO J 21:5955–5959
Wanke S, Samain MS, Vanderschaeve L, Mathieu G, Goetghebeur P, Neinhuis C (2006) Phylogeny of the genus Peperomia (Piperaceae) inferred from the trnK/matK region (cpDNA). Plant Biol (Stuttg) 8:91–102
Wijsman HJW (1982) On the inter-relationships of certain species of Petunia I. Taxonomic notes on the parental species of Petunia hybrida. Acta Bot Neerl 31:477–490
Wijsman HJW (1990) On the inter-relationships of certain species of Petunia VI. New names for the species of Calibrachoa formerly included into Petunia (Solanaceae). Acta Bot Neerl 39:101–102
Young ND, Healy J (2003) GapCoder automates the use of indel characters in phylogenetic analysis. BMC Bioinformatics 4:6
Zhu S, Fushimi H, Cai SQ, Komatsu K (2003) Phylogenetic relationship in genus Panax: inferred from chloroplast trnK gene and nuclear 18S rRNA gene sequences. Planta Med 69:647–653
Acknowledgments
We thank Mr Tsuguyoshi Aoki of Buenos Aires, Argentina, Mr Sebastião T. Nagase, Mr Nobuyuki Hiranaka, Mr Tomio Koshizawa, Mr Hideo Ohkubo, and Mr Roberto H. Ohkubo of São Paulo, Brazil, and Mr Masao Udagawa of Montevideo, Uruguay, for help in surveying the natural habitat.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
10265_2006_70_MOESM1_ESM.pdf
Supplement Fig. 1. Strict consensus tree of the 321 most parsimonious trees calculated from combined data of Hf1 exon and trnK intron sequences. Numbers below branches are bootstrap support values (1000 replicates). (PDF kb)
Rights and permissions
About this article
Cite this article
Chen, S., Matsubara, K., Omori, T. et al. Phylogenetic analysis of the genus Petunia (Solanaceae) based on the sequence of the Hf1 gene. J Plant Res 120, 385–397 (2007). https://doi.org/10.1007/s10265-006-0070-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-006-0070-z