Abstract
Key message
A total of seven glutathione reductase (GR) genes were identified in Triticum aestivum, which were used for comparative structural characterization, phylogenetic analysis and expression profiling with the GR genes of other cereal plants. The modulated gene expression and enzyme activity revealed the role of GRs in abiotic stress response in T. aestivum.
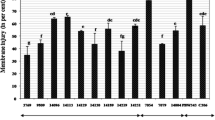
Glutathione reductase (GR) is an enzymatic antioxidant that converts oxidized glutathione (GSSG) into reduced glutathione (GSH) through the ascorbate–glutathione cycle. In this study, a total of seven GR genes forming two homeologous groups were identified in the allohexaploid genome of bread wheat (Triticum aestivum). Besides, we identified three GR genes in each Aegilops tauschii, Brachypodium distachyon, Triticum urartu and Sorghum bicolor, which were used for comparative characterization. Phylogenetic analysis revealed the clustering of GR proteins into two groups; class I and class II, which were predicted to be localized in cytoplasm and chloroplast, respectively. The exon–intron and conserved motif patterns were almost conserved in each group, in which a maximum of 10 and 17 exons were present in chloroplastic and cytoplasmic GRs, respectively. The protein structure analysis confirmed the occurrence of conserved pyridine nucleotide disulfide oxidoreductase (Pyr_redox) and pyridine nucleotide disulfide oxidoreductase dimerization (Pyr_redox_dim) domains in each GR. The active site of GR proteins consisted of two conserved cysteine residues separated by four amino acid residues. Promoter analysis revealed the occurrence of growth and stress-related cis-active elements. Tissue-specific expression profiling suggested the involvement of GRs in both vegetative and reproductive tissue development in various plants. The differential expression of TaGR genes and enhanced GR enzyme activity suggested their roles under drought, heat, salt and arsenic stress. Interaction of GRs with other proteins and chemical compounds of the ascorbate–glutathione cycle revealed their coordinated functioning. The current study will provide a foundation for the validation of the precise role of each GR gene in future studies.








Similar content being viewed by others
Data availability
All the data is available in the manuscript. The RNA seq data used in the study were freely available at Expression Atlas, URGI and NCBI databases.
References
Abhinandan K, Skori L, Stanic M, Hickerson NMN, Jamshed M, Samuel MA (2018) Abiotic stress signaling in wheat—an inclusive overview of hormonal interactions during abiotic stress responses in wheat. Front Plant Sci 9:734. https://doi.org/10.3389/fpls.2018.00734
Achary VMM, Reddy CS, Pandey P, Islam T, Kaul T, Reddy MK (2015) Glutathione reductase a unique enzyme: molecular cloning, expression and biochemical characterization from the stress adapted C 4 plant, Pennisetum glaucum (L.) R. Br. Mol Biol Rep 42(5):947–962. https://doi.org/10.1007/s11033-014-3832-z
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37(suppl_2):W202–W208. https://doi.org/10.1093/nar/gkp335
Barah P, Jayavelu ND, Rasmussen S, Nielsen HB, Mundy J, Bones AM (2013) Genome-scale cold stress response regulatory networks in ten Arabidopsis thaliana ecotypes. BMC Genomics 14:722. https://doi.org/10.1016/j.ijbiomac.2020.03.021
Bashir K, Nagasaka S, Itai RN, Kobayashi T, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2007) Expression and enzyme activity of glutathione reductase is upregulated by Fe-deficiency in graminaceous plants. Plant Mol Biol 65(3):277–284. https://doi.org/10.1007/s11103-007-9216-1
Bastian M, Heymann S, Jacomy M (2009) Gephi: an open-source software for exploring and manipulating networks visualization and exploration of large graphs. Proc. Third Int. ICWSM Conf. 3
Carillo P, Annunziata MG, Pontecorvo G, Fuggi A, Woodrow P, (2011) Salinity Stress and Salt Tolerance, Abiotic Stress in Plants - Mechanisms and Adaptations, Arun Shanker and B. Venkateswarlu, IntechOpen. https://doi.org/10.5772/22331
Chao YY, Hsu YT, Kao CH (2009) Involvement of glutathione in heat shock–and hydrogen peroxide–induced cadmium tolerance of rice (Oryza sativa L.) seedlings. Plant Soil 318(1–2):37. https://doi.org/10.1007/s11104-008-9815-x
Choulet F, Albert A, Theil S, Glover N, Barbe V, Daron J, Pingault L et al (2014) Structural and functional partitioning of bread wheat chromosome 3B. Science 18(345):1249721–1249721. https://doi.org/10.1126/science
Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008:1–12. https://doi.org/10.1155/2008/619832
Connell JP, Mullet JE (1986) Pea chloroplast glutathione reductase: purification and characterization. Plant Physiol 82(2):351–356. https://doi.org/10.1104/pp.82.2.351
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16(22):10881–10890. https://doi.org/10.1093/nar/16.22.10881
Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol 11(1):1–14. https://doi.org/10.1186/1471-2229-11-163
Dat J, Vandenabeele S, Vranova EVMM, Van Montagu M, Inzé D, Van Breusegem F (2000) Dual action of the active oxygen species during plant stress responses. CMLS 57(5):779–795. https://doi.org/10.1007/s000180050041
Deponte M (2013) Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. BiochimBiophys Acta Gen Subj 1830(5):3217–3266. https://doi.org/10.1016/j.bbagen.2012.09.018
Ding S, Wang L, Yang Z, Lu Q, Wen X, Lu C (2016) Decreased glutathione reductase2 leads to early leaf senescence in Arabidopsis. J Integr Plant Biol 58(1):29–47. https://doi.org/10.1111/jipb.12371
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797. https://doi.org/10.1093/nar/gkh340
Edwards EA, Rawsthorne S, Mullineaux PM (1990) Subcellular distribution of multiple forms of glutathione reductase in leaves of pea (Pisum sativum L.). Planta 180(2):278–284. https://doi.org/10.1007/BF00194008
El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL (2019) The Pfam protein families database in 2019. Nucleic Acids Res 47(D1):D427–D432
Emanuelsson O, Nielsen H, Heijne GV (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8(5):978–984. https://doi.org/10.1110/ps.8.5.978
Farooq MA, Li L, Ali B, Gill RA, Wang J, Ali S, Gill MB, Zhou W (2015) Oxidative injury and antioxidant enzymes regulation in arsenic-exposed seedlings of four Brassica napus L cultivars. Environ Sci Pollut Res 22(14):10699–10712. https://doi.org/10.1007/s11356-015-4269-1
Forman HJ, Zhang H, Rinna A (2009) Glutathione: overview of its protective roles, measurement, and biosynthesis. Mol Aspects Med 30(1–2):1–12. https://doi.org/10.1016/j.mam.2008.08.006
Foyer C, Lelandais M, Galap C, Kunert KJ (1991) Effects of elevated cytosolic glutathione reductase activity on the cellular glutathione pool and photosynthesis in leaves under normal and stress conditions. Plant Physiol 97(3):863–872. https://doi.org/10.1104/pp.97.3.863
Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. The proteomics protocols handbook. Humana press, Totowa, pp 571–607
Gholampour-Faroji N, Farazmand R, Hemmat J, Haddad-Mashadrizeh A (2019) Modeling, stability and the activity assessment of glutathione reductase from Streptococcus Thermophilus; insights from the in-silico simulation study. Comput Biol Chem 83:107121. https://doi.org/10.1016/j.compbiolchem.2019.107121
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48(12):909–930. https://doi.org/10.1016/j.plaphy.2010.08.016
Gill SS, Anjum NA, Hasanuzzaman M, Gill R, Trivedi DK, Ahmad I, Pereira E, Tuteja N (2013) Glutathione and glutathione reductase: a boon in disguise for plant abiotic stress defense operations. Plant Physiol Biochem 70:204–212. https://doi.org/10.1016/j.plaphy.2013.05.032
Golldack D, Lüking I, Yang O (2011) Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep 30(8):1383–1391. https://doi.org/10.1007/s00299-011-1068-0
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8(8):1494–1512. https://doi.org/10.1038/nprot.2013.084
Halliwell B, Foyer CH (1978) Properties and physiological function of a glutathione reductase purified from spinach leaves by affinity chromatography. Planta 139(1):9–17. https://doi.org/10.1007/BF00390803
Hanukoglu I (2015) Proteopedia: Rossmann fold: a beta-alpha-beta fold at dinucleotide binding sites. Biochem Mol Biol Educ 43(3):206–209. https://doi.org/10.1002/bmb.20849
Harshavardhan VT, Wu TM, Hong CY (2017) Glutathione reductase and abiotic stress tolerance in plants. In Glutathione in plant growth, development, and stress tolerance. Springer, Cham, pp 265–286. https://doi.org/10.1007/978-3-319-66682-2_12
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35(suppl_2):W585–W587. https://doi.org/10.1093/nar/gkm259
Hu X, Liu R, Li Y, Wang W, Tai F, Xue R, Li C (2010) Heat shock protein 70 regulates the abscisic acid-induced antioxidant response of maize to combined drought and heat stress. Plant Growth Regul 60(3):225–235. https://doi.org/10.1007/s10725-009-9436-2
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Huang C, He W, Guo J, Chang X, Su P, Zhang L (2005) Increased sensitivity to salt stress in an ascorbate-deficient Arabidopsis mutant. J Exp Bot 56(422):3041–3049
Huo N, Zhang S, Zhu T, Dong L, Wang Y, Mohr T, Hu T, Liu Z, Dvorak J, Luo M-C, Wang D, Lee J-Y, Altenbach S, Gu YQ (2018) Gene Duplication and evolution dynamics in the Homeologous regions Harboring multiple Prolamin and resistance Gene families in Hexaploid Wheat. Front Plant Sci 9:673
Islam S, Rahman IA, Islam T, Ghosh A (2017) Genome-wide identification and expression analysis of glutathione S-transferase gene family in tomato: gaining an insight to their physiological and stress-specific roles. PLoS One 12(11):e0187504. https://doi.org/10.1371/journal.pone.0187504
Jiang M, Zhang J (2002) Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J Exp Bot 53(379):2401–2410
Kajla M, Yadav VK, Khokhar J, Singh S, Chhokar RS, Meena RP, Sharma RK (2015) Increase in wheat production through management of abiotic stresses. JANS 7(2):1070–1080
Kaminaka H, Morita S, Nakajima M, Masumura T, Tanaka K (1998) Gene cloning and expression of cytosolic glutathione reductaserice (Oryza sativa L.). Plant Cell Physiol. 39(12):1269–1280. https://doi.org/10.1093/oxfordjournals.pcp.a029330
Kanwar MK, Bhardwaj R (2015) Arsenic induced modulation of antioxidative defense system and brassinosteroids in Brassica juncea L. Ecotoxicol Environ Saf 115:119–125
Karplus PA, Schulz GE (1987) Refined structure of glutathione reductase at 1.54 Å resolution. J Mol Biol 195(3):701–729. https://doi.org/10.1016/0022-2836(87)90191-4
Kaur A, Taneja M, Tyagi S, Sharma A, Singh K, Upadhyay SK (2020) Genome-wide characterization and expression analysis suggested diverse functions of the mechanosensitive channel of small conductance-like (MSL) genes in cereal crops. Sci Rep 10, 16583. https://doi.org/10.1038/s41598-020-73627-7
Kubo A, Sano T, Saji H, Tanaka K, Kondo N, Tanaka K (1993) Primary structure and properties of glutathione reductase from Arabidopsis thaliana. Plant Cell Physiol 34(8):1259–1266. https://doi.org/10.1093/oxfordjournals.pcp.a078548
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Lascano HR, Gomez LD, Casano LM, Trippi VS (1999) Wheat chloroplastic glutathione reductase activity is regulated by the combined effect of pH NADPH and GSSG Plant. Cell Physiol 40(7):683–690. https://doi.org/10.1093/oxfordjournals.pcp.a029593
Lee YP, Kim SH, Bang JW, Lee HS, Kwak SS, Kwon SY (2007) Enhanced tolerance to oxidative stress in transgenic tobacco plants expressing three antioxidant enzymes in chloroplasts. Plant Cell Rep. 26(5):591–598. https://doi.org/10.1007/s00299-006-0253-z
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327. https://doi.org/10.1093/nar/30.1.325
Liu Y, He C (2016) Regulation of plant reactive oxygen species (ROS) in stress responses: learning from AtRBOHD. Plant Cell Rep 35(5):995–1007
Liu Z, Xin M, Qin J, Peng H, Ni Z, Yao Y, Sun Q (2015) Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 15:152. https://doi.org/10.1186/s12870-015-0511-8
Liu HJ, Wang X, Yang ZL, Ren LL, Qian TT (2020) Identification and biochemical characterization of the glutathione reductase family from Populus trichocarpa. Plant Sci 294:110459. https://doi.org/10.1016/j.plantsci.2020.110459
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Madamanchi NR, Anderson JV, Alscher RG, Cramer CL, Hess JL (1992) Purification of multiple forms of glutathione reductase from pea (Pisum sativum L.) seedlings and enzyme levels in ozone-fumigated pea leaves. Plant Physiol 100(1):138–145. https://doi.org/10.1104/pp.100.1.138
Mahan JR, Burke JJ (1987) Purification and characterization of glutathione reductase from corn mesophyll chloroplasts. Physiol Plant 71(3):352–358. https://doi.org/10.1111/j.1399-3054.1987.tb04355.x
Malik WA, Wang X, Wang X, Shu N, Cui R, Chen X, Wang D, Lu X, Yin Z, Wang J, Ye W (2020) Genome-wide expression analysis suggests glutaredoxin genes response to various stresses in cotton. Int J Biol Macromol 153:470–491. https://doi.org/10.1016/j.ijbiomac.2020.03.021
Mande SS, Sarfaty S, Allen MD, Perham RN, Hol WG (1996) Protein–protein interactions in the pyruvate dehydrogenase multienzyme complex: dihydrolipoamide dehydrogenase complexed with the binding domain of dihydrolipoamide acetyltransferase. Structure 4(3):277–286. https://doi.org/10.1016/s0969-2126(96)00032-9
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43(D1):D222–D226. https://doi.org/10.1093/nar/gku1221
Marty L, Siala W, Schwarzländer M, Fricker MD, Wirtz M, Sweetlove LJ, Meyer Y, MeyerAJ RJP, Hell R (2009) The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. PNAS 106(22):9109–9114. https://doi.org/10.1073/pnas.0900206106
Molina A, Bueno P, Marín MC, Rodríguez-Rosales MP, Belver A, Venema K, Donaire JP (2002) Involvement of endogenous salicylic acid content, lipoxygenase and antioxidant enzyme activities in the response of tomato cell suspension cultures to NaCl. New Phytol 156(3):409–415
Mullineaux PM (1997) Glutathione reductase: regulation and role in oxidative stress. Oxidative stress and the molecular biology of antioxidant defence 667–713
Nussbaumer T, Martis MM, Roessner SK, Pfeifer M, Bader KC, Sharma S, Gundlach H, Spannagl M (2012) MIPS PlantsDB: a database framework for comparative plant genome research. Nucleic Acids Res 41(D1):D1144–D1151. https://doi.org/10.1093/nar/gks1153
Pandey S, Subramanaym Reddy C, Yaqoob U, Negi YK, Arora S (2015) Insilico analysis of cis acting regulatory elements CAREs in upstream regions of Ascorbate Glutathione Pathway Genes from Oryza sativa. BiochemPhysiol 4(159):2. https://doi.org/10.4172/2168-9652.1000159
Papatheodorou I, Fonseca NA, Keays M, Tang YA, Barrera E, Bazant W, Burke M, Füllgrabe A, Fuentes AMP, George N, Huerta L (2018) Expression Atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res 46(D1):D246–D251. https://doi.org/10.1093/nar/gkx1158
Pierleoni A, Martelli PL, Fariselli P, Casadio R (2006) BaCelLo: a balanced subcellular localization predictor. Bioinformatics 22(14):e408–e416. https://doi.org/10.1093/bioinformatics/btl222
Pingault L, Choulet F, Alberti A, Glover N, Wincker P, Feuillet C, Paux E (2015) Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome. Genome Biol 16(1):29. https://doi.org/10.1186/s13059-015-0601-9
Rao AC, Reddy AR (2008) Glutathione reductase: a putative redox regulatory system in plant cells. Sulfur assimilation and abiotic stress in plants. Springer, Berlin, pp 111–147
Rao MV, Paliyath G, Ormrod DP (1996) Ultraviolet-B and ozoneinducedbiochemical changes in antioxidant enzymes of Arabidopsis thaliana. Plant Physiol 110:125–136. https://doi.org/10.1104/pp.110.1.125
Rescigno M, Perham RN (1994) Structure of the NADPH-binding motif of glutathione reductase: efficiency determined by evolution. Biochemistry 33(19):5721–5727. https://doi.org/10.1021/bi00185a008
Sairam RK, Srivastava GC, Agarwal S, Meena RC (2005) Differences in antioxidant activity in response to salinity stress in tolerant and susceptible wheat genotypes. Biol Plantarum 49(1):85
Sakuma YK, Maryyama F, Qin Y, Osakabe K, Shinozaki KY, Yamaguchi S (2006) Dual function of an Arabidopsis transcription factor dreb2a in water-stress-responsive and heat-stress-responsive gene expression. PNAS 103:18822–18827. https://doi.org/10.1073/pnas.0605639103
Scandalios JG (2005) Oxidative stress: molecular perception and transduction of signals triggering antioxidant gene defenses. BJMBR 38(7):995–1014. https://doi.org/10.1590/S0100-879X2005000700003
Seo J, Gordish-Dressmanm H, Hoffman EP (2006) An interactive power analysis toolfor microarray hypothesis testing and generation. Bioinformatics 22:808–814
Sharma H, Taneja M, Upadhyay SK (2020) Identification, characterization and expression profiling of cation-proton antiporter superfamily in Triticum aestivum L. and functional analysis of TaNHX4-B. Genomics 112(1):356–370. https://doi.org/10.1016/j.ygeno.2019.02.015
Sharma A, Sharma H, Upadhyay SK (2020) Thaumatin-like proteins: Molecular characterization, evolutionary analysis and expression profiling in major cereal crops. Plant Sci 290:110317. https://doi.org/10.1101/2020.09.24.311928
Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signalling pathways. CurrOpin Plant Biol 3:217–223. https://doi.org/10.1016/S1369-5266(00)80068-0
Shumayla, Sharma S, Pandey AK, Singh K, Upadhyay SK (2016) Molecular characterization and global expression analysis of lectin receptor kinases in bread wheat (Triticum aestivum). PLoS One 11(4):e0153925. https://doi.org/10.1371/journal.pone.0153925
Shumayla, Tyagi S, Sharma A, Singh K, Upadhyay SK (2019) Genomic dissection and transcriptional profiling of Cysteine-rich receptor-like kinases in five cereals and functional characterization of TaCRK68-A. Int J Biol Macromol 134:316–329. https://doi.org/10.1016/j.ijbiomac.2019.05.016
Smirnoff N, Colombé SV (1988) Drought influences the activity of enzymes of the chloroplast hydrogen peroxide scavenging system. J Exp Bot 39(8):1097–1108
Souri Z, Karimi N, de Oliveira LM (2018) Antioxidant enzymes responses in shoots of arsenic hyperaccumulator, IsatiscappadocicaDesv., under interaction of arsenate and phosphate. Environ Technol 39(10):1316–1327
Stevens RG, Creissen GP, Mullineaux PM (1997) Cloning and characterisation of a cytosolic glutathione reductase cDNA from pea (Pisum sativum L.) and its expression in response to stress. Plant Mol Biol 35(5):641–54
Szklarczyk D, Santos A, Von Mering C, Jensen LJ, Bork P, Kuhn M (2016) STITCH 5: augmenting protein—chemical interaction networks with tissue and affinity data. Nucleic Acids Res 44(D1):D380–D384. https://doi.org/10.1093/nar/gkv1277
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613. https://doi.org/10.1093/nar/gky1131
Tahmasebi A, Aram F, Ebrahimi M, Mohammadi-Dehcheshmeh M, Ebrahimie E (2012) Genome-wide analysis of cytosolic and chloroplastic isoforms of glutathione reductase in plant cells. Plant Omics 5(2):94–102
Takeda T, Ishikawa T, Shigeoka S, Hirayama O, Mitsunaga T (1993) Purification and characterization of glutathione reductase from Chlamydomonas reinhardtii. Microbios 139(9):2233–2238
Trivedi DK, Gill SS, Yadav S, Tuteja N (2013) Genome-wide analysis of glutathione reductase (GR) genes from rice and Arabidopsis. Plant Signal Behav 8(2):e23021. https://doi.org/10.4161/psb.23021
Tyagi S, Sharma S, Taneja M, Kumar R, Sembi JK, Upadhyay SK (2017) Superoxide dismutases in bread wheat (Triticum aestivum L.): comprehensive characterization and expression analysis during development and biotic and abiotic stresses. Agri Gene 6:1–13
Tyagi S, Sembi JK, Upadhyay SK (2018) Gene architecture and expression analyses provide insights into the role of glutathione peroxidases (GPXs) in bread wheat (Triticum aestivum L.). J Plant Physiol 223:19–31. https://doi.org/10.1016/j.jplph.2018.02.006
Tyagi S, Shumayla VPC, Singh K, Upadhyay SK (2020) Molecular characterization of ascorbate peroxidase (APX) and APX-related (APX-R) genes in Triticum aestivum L. Genomics 112:4208–4223. https://doi.org/10.1016/j.ygeno.2020.07.023
Tyagi S, Shumayla SK, Upadhyay SK (2021) Molecular characterization revealed the role of catalases under abiotic and arsenic stress in bread wheat (Triticum aestivum L). J Hazard Mater 403:123585. https://doi.org/10.1016/j.jhazmat.2020.123585
Verma PK, Verma S, Pande V, Mallick S, Tripathi R, Dhankher OP, Chakrabarty D (2016) Overexpression of rice glutaredoxin OsGrx_C7 and OsGrx_C2. 1 reduces intracellular arsenic accumulation and increases tolerance in Arabidopsis thaliana. Front Plant Sci 7:740. https://doi.org/10.3389/fpls.2016.00740
Wang X, Cai J, Liu F, Dai T, Cao W, Wollenweber B, Jiang D (2014) Multiple heat priming enhances thermo-tolerance to a later high temperature stress via improving subcellular antioxidant activities in wheat seedlings. Plant PhysiolBiochem 74:185–192. https://doi.org/10.1016/j.plaphy.2013.11.014
Wittkopp PJ, Kalay G (2012) Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet 13(1):59–66
Xiong J, (2006) Protein motifs and domain prediction. Essential bioinformatics, Cambridge University Press, 85–94
Xu G, Guo C, Shan H, Kong H (2012) Divergence of duplicate genes in exon–intron structure. PNAS 109(4):1187–1192. https://doi.org/10.1073/pnas.1109047109
Yan L, Liu ZQ, Xu YH, Lu K, Wang XF et al (2013) Auto- and cross-repression of three Arabidopsis WRKY transcription factors WRKY18, WRKY40 and WRKY60 negatively involved in ABA signalling. J Plant Growth Regul 32:399–416
Yang J, Zhang Y (2015) Protein structure and function prediction using I-TASSER. Curr Protoc Bioinformatics 52(1):5–8
Yu CS, Cheng CW, Su WC, Chang KC, Huang SW, Hwang JK, Lu CH (2014) CELLO2GO: a web server for protein subCELlularLOcalization prediction with functional gene ontology annotation. PLoS One 9(6):e99368. https://doi.org/10.1371/journal.pone.0099368
Zhang Y, Bond CS, Bailey S, Cunningham ML, Fairlamb AH, Hunter WN (1996) The crystal structure of trypanothione reductase from the human pathogen Trypanosoma cruzi at 2.3 Å resolution. Protein Sci 5(1):52–61. https://doi.org/10.1002/pro.5560050107
Zhang Y, Liu Z, Khan AA, Lin Q, Han Y, Mu P, Liu Y, Zhang H, Li L, Meng X, Ni Z (2016) Expression partitioning of homeologs and tandem duplications contribute to salt tolerance in wheat (Triticum aestivum L.). Sci Rep 6:21476. https://doi.org/10.1038/srep21476
Acknowledgements
The authors are grateful to the Panjab University, Chandigarh for research facilities, Ensembl Plants, URGI and NCBI for data availability. SKU is grateful to the Council of Scientific and Industrial Research (CSIR) for a research grant (No. 38(1489)/19/EMR-II). M is grateful to UGC for JRF. AK, ST and S are grateful to CSIR for SRF and RA, respectively.
Funding
Department of Science and Technology (DST), Government of India, provided partial financial support under the Promotion of University Research and Scientific Excellence (PURSE) grant scheme.
Author information
Authors and Affiliations
Contributions
SKU conceived the idea. M and SKU designed the experiments. M, AK and S performed the experiments. M, ST, S, KS and SKU analyzed the data. M and SKU wrote the manuscript. All the authors have read and finalized the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Consent for publication
All the authors have approved the manuscript for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
299_2021_2717_MOESM1_ESM.xlsx
Supplementary file1 Supplementary Table S1 List of identified GR genes with their proposed nomenclature in Aegilops tauschii, Brachypodiumdistachyon, Sorhum bicolor, Triticum aestivum and Triticum urartu. (XLSX 10 KB)
299_2021_2717_MOESM2_ESM.xlsx
Supplementary file2 Supplementary Table S2 Characterization table of GR genes and proteins of Aegilops tauschii, Brachypodiumdistachyon, Sorhum bicolor, Triticum aestivum and Triticum urartu. (XLSX 11 KB)
299_2021_2717_MOESM3_ESM.xlsx
Supplementary file3 Supplementary Table S3 Domain organization of GR proteins in Aegilops tauschii, Brachypodium distachyon, Sorghum bicolor, Tritricum aestivum and Triticum urartu and motif sequences. (XLSX 12 KB)
299_2021_2717_MOESM4_ESM.xlsx
Supplementary file4 Supplementary Table S4 List of cis-regulatory elements identified in the promoter region of GR genes of Aegilops tauschii, Brachypodiumdistachyon, Sorghum bicolor, Tritricumaestivum and Triticum urartu. (XLSX 11 KB)
299_2021_2717_MOESM5_ESM.xlsx
Supplementary file5 Supplementary Table S5 Annotation and abbreviation of interacting proteins of Brachypodium distachyon, Sorghum bicolor and Tritricum aestivum. (XLSX 11 KB)
299_2021_2717_MOESM6_ESM.xlsx
Supplementary file6 Supplementary Table S6 List of qRT-PCR primers. Annotation and abbreviation of interacting compounds with GR of Brachypodiumdistachyon, Sorghum bicolor and Tritricum aestivum. (XLSX 10 KB)
299_2021_2717_MOESM8_ESM.jpg
Supplementary file8 Supplementary Fig. 1. Figure shows the correlation analysis between replicates of expression data in A tissue developmental stages B stress conditions. (XLSX 33 KB)
Rights and permissions
About this article
Cite this article
Madhu, Kaur, A., Tyagi, S. et al. Exploration of glutathione reductase for abiotic stress response in bread wheat (Triticum aestivum L.). Plant Cell Rep 41, 639–654 (2022). https://doi.org/10.1007/s00299-021-02717-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-021-02717-1