Abstract
Mycological (mycotoxigenic Fusarium and aflatoxigenic Aspergillus spp.) and multiple mycotoxins [aflatoxin B1 (AFB1), fumonisin B (FB), deoxynivalenol and zearalenone] surveillance was conducted on raw whole grain sorghum (Sorghum bicolor) and pearl millet (Pennisetum glaucum) produced on smallholder farms, and processed products sold at open markets in northern Namibia. Fungal contamination was determined with morphological methods as well as with quantitative Real-Time PCR (qPCR). The concentrations of multiple mycotoxins in samples were determined with liquid chromatography tandem mass spectrometry. The incidence of mycotoxigenic Fusarium spp., Aspergillus flavus and A. parasiticus, as well as the concentrations of AFB1 and FB were significantly (P < 0.001) higher in the malts as compared to the raw whole grains, with Aspergillus spp. and AFB1 exhibiting the highest contamination (P < 0.001). None of the analysed mycotoxins were detected in the raw whole grains. Aflatoxin B1 above the regulatory maximum level set by the European Commission was detected in sorghum (2 of 10 samples; 20%; 3–11 µg/kg) and pearl millet (6 of 11 samples; 55%; 4–14 µg/kg) malts. Low levels of FB1 (6 of 10 samples; 60%; 15–245 µg/kg) were detected in sorghum malts and no FB was detected in pearl millet malts. Contamination possibly occurred postharvest, during storage, and/or transportation and processing. By critically monitoring the complete production process, the sources of contamination and critical control points could be identified and managed. Mycotoxin awareness and sustainable education will contribute to reducing mycotoxin contamination. This could ultimately contribute to food safety and security in northern Namibia where communities are exposed to carcinogenic mycotoxins in their staple diet.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sorghum (Sorghum bicolor) and pearl millet (Pennisetum glaucum) constitute half of the total cereal crop production in Africa and play an important role in the maintenance of food security [1]. Their high yield rate and adaptation to extreme environmental conditions make them suitable for agricultural utilization in most regions in Africa. As a food commodity, sorghum and pearl millet are commonly ground into flour or malted and used to prepare food and beverages, including weaning foods [1,2,3]. In Africa, sorghum and millet are largely subsistence food crops, but is increasingly forming the foundation of successful food and beverage industries [1]. Subsistence crops are cultivated for home consumption, i.e., for food preparation and beer brewing, as well as for informal trading.
Contamination of staple grains with mycotoxigenic fungi and mycotoxins occurs in many regions of the world, impacting negatively on food security, crop quality and trade [4, 5]. Mycotoxigenic fungi can infiltrate deep into sorghum and pearl millet matrices and produce mycotoxins during the pre-harvest, storage, transportation, processing, and marketing stages. Toxicologically significant mycotoxins include aflatoxin B1 (AFB1) produced by Aspergillus spp., and fumonisin B1 (FB1), fumonisin B2 (FB2), fumonisin B3 (FB3), deoxynivalenol (DON) and zearalenone (ZEA) produced by Fusarium spp. [3, 4]. These toxins cause a variety of biochemical effects, including carcinogenic, mutagenic, teratogenic, estrogenic, neurotoxic, hepatotoxic, nephrotoxic, cytotoxic, and immunosuppressive conditions [6]. AFB1 is mainly produced by A. flavus and A. parasiticus. After ingestion, AFB1 forms DNA adducts which initiates carcinogenesis and can work synergistically with hepatitis B virus [7]. It poses a serious threat to human and animal health by causing hepatotoxicity, teratogenicity, immunotoxicity as well as liver cancer, and is classified a Group 1 carcinogen by the International Agency for Research on Cancer (IARC) [8, 9]. The fumonisins are mainly produced by F. verticillioides and F. proliferatum. FB causes a decrease in complex sphingolipids, glycerophospholipids, and cholesterol, which are essential for cell membrane integrity, resulting in possible intestinal epithelial cell proliferation, disruption of cytokine production and modulation of intestinal barrier function [10, 11]. FB has been associated with neural tube defects, stunting in children and oesophageal cancer, and is classified a Group 2B carcinogen [8]. DON, a vomitoxin, is produced by fungi belonging to the F. graminearum spp. complex [12]. It causes intestinal barrier impairment and immunostimulatory effects in low doses in animals and emesis, reduction in feed conversion rate, and immunosuppression in high doses. Contamination by ZEA is mainly caused by F. graminearum, F. equiseti, F. culmorum, F. cerealis and F. semitectum. ZEA is an estrogenic mycotoxin affecting male and female reproductive systems [13]. Chronic exposure to mycotoxins, such as DON or DON and FB in combination, has been suggested to modulate child growth by inducing a poor appetite, gut impairment, inflammatory diarrhoea, decreased nutrient absorption and systemic immune activation [10, 14]. Co-exposure to multiple mycotoxins can have additive effects, contributing to existing health conditions and disease burden [10].
Previous reviews on mycotoxin contamination of sorghum and millet in Africa concentrated primarily on Central, Eastern and Western Africa [15]. Co-occurrence of AFs and FBs has been documented in sorghum and pearl millet from smallholder farmers under the direction of the International Institute for Tropical Agriculture, Nigeria. A surveillance study evaluating the levels of multiple mycotoxins in sorghum from Burkina Faso, Ethiopia, Mali, and Sudan resulted in 33% of 1533 samples contaminated with at least one of the AFs and FBs, sterigmatocystin, Alternaria toxins, ochratoxin A and ZEA [16]. Only a few reports have been documented on sorghum and millet in South Africa [17, 18], Botswana, Lesotho, Malawi, Mozambique, Zambia, and Zimbabwe [19]. Several Fusarium spp. have been isolated from whole grain sorghum in South Africa with Fusarium verticillioides, F. proliferatum and the F. graminearum spp. complex representing the main mycotoxigenic fungi [18]. Data indicated that the contamination levels of Fusarium and Aspergillus spp. and their mycotoxins do not pose a threat to the production of commercial sorghum in South Africa [17]. F. verticillioides and F. nygamai are the main fungi contaminating sorghum and millet in Lesotho and Zimbabwe [20]. In Gaborone, Botswana, 46 traditional sorghum malt, wort, and beer samples were collected from villages [21]. F. verticillioides contamination was detected in 63% samples and Aspergillus flavus in 37%. AFs were not detected, whilst FB1 was detected in three malt samples (47–1316 µg/kg), and ZEA in malt (102–2213 µg/kg), wort (26–285 µg/L) and beer (20–201 µg/L) samples, respectively. There are no reports available on the occurrence of fungi and mycotoxins in raw whole grain sorghum and pearl millet in Namibia, contamination that occurs during storage, and limited reports on processed grains. In Namibia, mycotoxins below the regulatory level of the European Union (EU) were detected in pearl millet meals [3], while AFB1 above the EU regulatory level was detected in sorghum malts used for brewing of oshikundu [3], omalodu and otombo [22]. There remain a huge knowledge gap concerning fungal and mycotoxin contamination in raw staple grains produced by smallholder farmers in Namibia, and the effects of storage and processing.
Many countries have instituted risk management practices by setting regulatory maximum levels (MLs) for mycotoxins in unprocessed staple grains intended for direct human consumption, and for processed grains [5]. AFB1, FB1 and FB2 are the most important mycotoxins contaminating staple grains and are regulated worldwide [5]. Strict regulation of mycotoxin levels in food exist in high-income countries with high levels of food safety control to guard against the harmful effects on human health [5]. In low-income countries, mycotoxin regulations are often absent or partially implemented, leading to circumstances where mycotoxin exposures are high. In many African countries, there are large subsistence farming populations reliant on subsistence grains as their primary staple food, consuming relatively large amounts compared to urban societies [23]. Despite the reported high dietary levels of mycotoxins, legislation for their control is absent in most countries in southern Africa [19]. When last surveyed, only a few countries in Africa have mycotoxin regulations, which are primarily linked to aflatoxin exposure in the most common dietary staples (Table S1) [24, 25]. Populations that are worst affected include smallholder farmers, where mono-cereal crops are harvested and locally consumed [5]. Subsistence-grown grains can be heavily contaminated and, being consumed in large amounts, results in high exposures and consequently raises health concerns [5, 23]. Smallholder farmers in northern Namibia utilize undiversified diets due to drought conditions and are heavily reliant on sorghum and pearl millet as a staple food [3]. Relevant geographical areas include the Oshana (production of sorghum and pearl millet) and Kavango (mainly production of pearl millet) regions. Locally, the raw and processed grains are sold at open markets.
The current study determined the incidence of mycotoxigenic Fusarium and aflatoxigenic Aspergillus spp., and the levels of multiple mycotoxins (AFB1, FB1, FB2, FB3, DON and ZEA) in raw whole grain sorghum and pearl millet collected from smallholder farms and processed products sold at local markets in the Oshana region of northern Namibia. Morphological as well as molecular techniques using species-specific primers and quantitative Real-time polymerase chain reaction (qPCR) were used to determine the incidence of the fungi. The concentrations of multiple mycotoxins in samples were determined with liquid chromatography tandem mass spectrometry (LC–MS/MS).
Materials and Methods
Field Study and Sampling of Sorghum and Pearl Millet
Raw whole grain sorghum and pearl millet samples (± 2 kg) were collected postharvest, prior to storage and processing, from 10 randomly selected smallholder farms in Oshakati in the Oshana region of northern Namibia during July 2018 (Table 1). Processed samples (malted sorghum and pearl millet) were obtained from 12 and 9 randomly selected vending stalls from the Oshakati and Ondangwa open markets, respectively (Table 1). The Oshakati smallholder communal farmers service both the Oshakati and Ondangwa open markets. Figure 1 depicts a geographical map of the smallholder farming sampling sites N1-N10 near Oshakati, as determined with GPS. Standardised sampling protocols adapted from “The Fusarium Laboratory Manual” [26] were followed. Labelling of the samples was done according to procedure described by Safrinet [27]. The first three letters in the sample code (NAM) denoted the locality. The numeric value in the sample code represented the number of the sampling site. The last letter denotes the substratum (“S” for sorghum and M” for pearl millet). Uncontaminated (control) sorghum and pearl millet were obtained from the Mycotoxin analysis laboratory of the Southern Africa Grain Laboratory (SAGL; South Africa). To confirm the purity of the control sorghum and pearl millet samples, they were plated out onto potato dextrose agar (PDA, Lasec Group Cat no. SKU MNCM0018) to determine fungal contamination and analyzed with LC–MS/MS to determine the presence of mycotoxins. The original grain samples were stored at 4 °C, while a 200 g subsample of each original was ground to a fine meal and stored at − 20 °C for molecular and mycotoxin analyses. Mycological analysis on the original samples was concluded within four months after collection.
Morphological Determination of the Incidence of Mycotoxigenic Fusarium and Aflatoxigenic Aspergillus spp. in Sorghum and Pearl Millet Samples
Isolation and Enumeration of the Fungi from Raw Whole Grain Samples
Fusarium spp.
Subsamples of whole grain sorghum and pearl millet kernels (100 g) were surface-sterilised and rinsed with sterile distilled water [28]. One hundred kernels from each sample were plated out, five kernels per Petri dish, on modified Malt extract agar (MEA, Merck Cat. no. 105398) containing 150 mg/L novobiocin (Sigma-Aldrich, Merck Cat. no. CDS020662) and incubated at 25 °C for 10–14 days. Fusarium spp. that developed from the kernels were identified according to their morphological characteristics [29, 30].
Aspergillus spp.
A subsample of kernels was surface sterilized, plated onto Aspergillus differentiation agar (AFPA, Merck Cat. no. 17121) and incubated at 30 °C for 5 days. The presence of a yellow-orange pigment visible at the bottom of the agar plates was used to determine the presence of A. flavus and/or A. parasiticus in samples [31].
Isolation and Enumeration of the Fungi in Processed Samples
Contamination of the samples with mycotoxigenic Fusarium and aflatoxigenic Aspergillus spp. was determined by the dilution plate method [26]. For detection of Fusarium spp., dilutions were plated out on Van Wyk’s Fusarium selective medium [32] and for A. flavus and A. parasiticus on AFPA. The number of colony forming units per gram sample (cfu/g sample) were recorded.
Molecular Identification and Quantification of Mycotoxigenic Fusarium and Aflatoxigenic Aspergillus spp. in Sorghum and Pearl Millet Samples
Fungal Reference Strains
Fusarium spp. reference strains were obtained from the Applied Microbial and Health Biotechnology Institute (AMHBI), Cape Peninsula University of Technology (CPUT), South Africa (F. verticillioides MRC 826, F. proliferatum MRC 8550 and F. graminearum MRC 6010). Aspergillus reference strains were obtained from AMHBI, CPUT [A. parasiticus (0200), A. flavus (0645 and 3954)] and from the American Type Culture Collection (ATCC, Virginia, USA) [A. parasiticus (CBS100926 AP1, CBS103.57 AP2 and CBS571.65 AP3) and A. flavus (CBS100927 AF1, CBS100.45 AF2 and CBS114062 AF3)].
Extraction of DNA from Fusarium and Aspergillus spp. Reference Cultures, Sorghum, and Pearl Millet Samples
Fungi were cultured in 100 ml Potato dextrose broth (PDB, Merck Cat. no. P6685) in 250 ml Erlenmeyer flasks at 26 °C on a rotary shaker for 14 days. The mycelium was harvested by filtration using a sterilized muslin cloth, and ground to a powder in liquid nitrogen with a mortar and pestle. Raw and processed sorghum and pearl millet samples (20 g) were ground to a powder using a laboratory mill (C and N laboratory mill, size 8, Christy and Norris Ltd. Engineers, Chelmsford, England). Prior to DNA extraction, the sample powder was further homogenized into a fine powder in liquid nitrogen using a mortar and pestle.
Total DNA was extracted from each sample (2 g) using a DNeasy Plant Mini Kit (Qiagen Cat. no. 69104) according to the procedure supplied by the manufacturer. The procedure was modified by the addition of 10 ml cetyltrimethylammonium bromide (CTAB)/polyvinyl-pyrrolidone (PVP) lysis buffer [33], 40 µl Proteinase K (10 mg/ml) and incubation for 2 h at 65 °C on a rotary shaker (200 rpm). The 500 µl flow through from the QIAshredder column in the DNeasy kit was transferred to a new 2 ml Eppendorf tube and an equal volume of phenol:chloroform:isoamylalcohol (P:C:I) (25:24:1; v/v) added, and the sample centrifuged at 14,000 rpm for 20 min. The top layer (450 µl) was transferred into a new 2 ml Eppendorf and an equal volume of chloroform/isoamylalcohol (24:1; v/v) (CI) added, mixed and centrifuge at 14,000 rpm for 20 min. This was done twice. The supernatant (350 µl) was transferred to a new 2 ml Eppendorf tube and continued with step 13 in the DNeasy kit protocol according to the manufacturer’s instructions. A Nanodrop 2000 Spectrophotometer (ThermoFisher Scientific, Inqaba Biotechnical Industries) was used to determine the DNA concentrations and purities by comparing the absorbance ratios A260/A280 and A260/A230. The quality of the DNA was visualized on a 0.8% agarose gel laced with ethidium bromide (5 µl) and run in 1 × Tris–acetate and EDTA (TAE) buffer electrophoresis at 70 V for 45 min [33]. A molecular weight marker, λ Hind III (ThermoFisher Scientific Cat. no. SM0101) (3 µl), containing a loading dye (2 µl), was included. Genomic DNA samples were diluted to a final concentration of 30 ng/µl and stored at − 20 °C until analysed with qPCR.
Quantitative Real-Time PCR
Primer Sequences
The species-specific primer sequences used for identification and quantification of Fusarium and Aspergillus spp. DNA are listed in Table S2. The Fusarium spp. primers were adapted from Nicolaisen et al. [34]. The primer design was based on the alignments of the elongation factor 1-alpha (EF1α) gene. The Aspergillus spp. primers were designed based on the sequence alignments of the internal transcribed spacer 2 (ITS2) region of the DNA of several strains from different origins, retrieved from nucleotide databases [35].
Optimization of qPCR Conditions
Quantitative Real-time PCR was performed with a CFX96 Real-time PCR detection system (Bio-Rad, California, USA). The optimum conditions for qPCR were determined by running trial and error experiments and modifying the conditions in the protocols by changing the annealing temperatures [34,35,36]. Optimization reactions were carried out separately on three reference strains of Fusarium spp. (F. verticillioides MRC 826, F. proliferatum MRC 8550 and F. graminearum MRC 6010) and on two Aspergillus spp. reference strains (A. parasiticus AP1 and A. flavus AF3) using control sorghum and pearl millet matrixes. DNA standard curves were prepared by diluting the respective fungal gDNA (2.5 ng/μl) in control sorghum and pearl millet gDNA (30 ng/μl), to obtain a four-fold dilution series. Standard curves, including a range of gDNA concentrations, were analysed to confirm acceptable ranges of the performance parameters [37].
Quantification of Mycotoxigenic Fusarium and Aflatoxigenic Aspergillus spp. DNA with qPCR
Quantitative Real-time PCR assays were carried out in triplicate in a total volume of 25 μl consisting of 12.5 μl 2 × SsoAdvanced universal SYBR Green Supermix (Bio-Rad Cat. no. 1725270), 250 nM of each primer and 2 μl template DNA (30 ng/μl). The assays were performed in 96-well plates using the protocols described in Table S3. Each 96-well plate included standard curve, negative control, and non-template control samples. The criteria for acceptance followed were according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) Guidelines [37]. Parameters included annealing temperatures, % efficiency, correlation coefficients (R2) and slopes (M). Quantification was conducted using the qBase + software (Bio-Rad), and the cycle threshold (Ct) values of each maize sample plotted against the logarithm of the known DNA concentrations starting quantity of standard template for each dilution to obtain the amount of fungal DNA concentration in the starting material.
Detection and Quantification of Multiple Mycotoxins with LC–MS/MS
Analytical Standards
Analytical standards of FB1, FB2 and FB3 (purity ≥ 97%) were obtained from the Mycotoxicology Research Group of the Institute of Biomedical and Microbial Biotechnology, CPUT. Analytical standards of AFB1 (Cat. no. A6636), ZEA (Cat. no. Z2125), and DON (Cat. no. D0156) were obtained from Sigma-Aldrich (Merck). Individual stock solutions (0.1 mg/ml) of AFB1 and ZEA were prepared in acetonitrile, while FB1, FB2 and FB3 and DON were prepared in acetonitrile-H2O (1:1). Working solutions in acetonitrile-H2O (1:1) containing (i) AFB1 and ZEA (200 ng/ml) individual concentrations, and (ii) FB1, FB2 and FB3 and DON (5 µg/ml) individual concentrations were prepared using aliquots of the stock solutions. Two-fold dilution series of the mycotoxin working solutions were prepared by utilising an extract prepared from control sorghum and pearl millet, respectively. The matrix-matched standards were used to compensate for matrix effects in the analysis.
Extraction Method
Multiple mycotoxins were extracted from raw whole grain and processed sorghum and pearl millet samples following the method described by Alberts et al. [38]. Extraction solvent [methanol: acetonitrile: water; (25:25:50); 100 ml] was added to ground samples (10 g) and the mixture placed on a rotary shaker for 30 min at 80 rpm. The extracts were centrifuged at 4000×g for 10 min at 4 °C in a refrigerated Sorvall RC-3B centrifuge (DuPont, Norwalk, Connecticut, USA). The supernatant was diluted (1:1) with methanol: water (25:75), filtered (Whatman No. 4 filter paper) and analysed with LC–MS/MS. Control sorghum and pearl millet samples, FAPAS (Cat no T22110QC; The Food and Environmental Research Agency, York, England; contains DON, ZEA and NIV) and Biopure (Cat no QCM3C2; Industrial Analytical, Kyalami, South Africa; contains FB1, FB2 and FB3) certified quality control samples, containing mycotoxins in the expected concentration ranges, were included in each run.
LC–MS/MS Analyses
The mycotoxins were separated on a reversed-phase BEH C18 column (2.1 × 50 mm; particle size 1.7 µm; Waters, Milford, MA, USA) and analysed with positive electrospray ionisation (ESI) in the multiple reaction monitoring (MRM) mode in a Waters Acquity Ultra Performance Liquid Chromatograph (UPLC) coupled to a Waters Xevo TQ tandem quadrupole mass spectrometer [38]. Eluent A was water and eluent B was acetonitrile, both containing 0.1% formic acid. The elution gradient consisted of an initial mobile phase composition (2% B) held constant for 0.5 min, followed by a linear gradient to 40% B within 7 min and to 70% B over 3 min, followed by a 1-min wash step at 100% B and finally a 3-min column re-equilibration to 2% B for a total run time of 15 min. The flow rate of the mobile phase was 0.35 ml/min. For each compound, one precursor and two product ions were monitored, one product ion for quantification and one for confirmation (Table S4). Quantification was conducted using The TargetLynx™ Application Manager of Masslynx 4.1 (Waters Corporation, Massachusetts, USA) for sample data acquisition, processing, and reporting for quantitative results relative to analytical standard calibrations. Validation experiments were performed to confirm the accuracy of the results, as described by the United States Food and Drug Administration [39]. Validation parameters included percentage (%) recovery of the extraction method, limit of quantification (LOQ), relative standard deviation for repeatability (RSDr) and the coefficient of determination (R2) of the respective mycotoxin calibration curves (Table 2).
Statistical Analyses
The NCSS Version 11 software [40] was used for statistical analysis. Data was analysed within a generalised linear model ANOVA. P < 0.05 was used as statistical significance. Correlation coefficients® were determined using R software, version 4.02 [41].
Results
Morphological Determination of the Incidence of Mycotoxigenic Fusarium and Aflatoxigenic Aspergillus spp. in Raw Whole Grain and Processed Sorghum and Pearl Millet Samples
Raw Whole Grain Samples
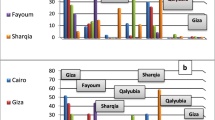
Samples NAM-1, NAM-2 and NAM-3 for both sorghum and pearl millet, obtained from smallholder farming sampling sites N1, N2 and N3 near Oshakati, exhibited the highest incidence of fungal contamination (P < 0.001) (Table 3). The contamination frequency of Fusarium spp. in sorghum was 80%, with NAM-1S (42%), NAM-2S (27%) and NAM-3S (39%) exhibiting the highest percentage kernel infection (P < 0.001). The Aspergillus spp. were not detected in sorghum samples, which confirmed results obtained during a study on commercial sorghum produced in South Africa [17]. In the current study, the contamination frequency of Fusarium spp. in pearl millet was 80%, with NAM-1M (9%), NAM-2M (4%) and NAM-3M (5%) exhibiting the highest percentage kernel infection (P < 0.05). The contamination frequency of the Aspergillus spp. in pearl millet samples was 33% with NAM-2M (10%), NAM-3M (10%) and NAM-8M (10%) exhibiting the highest percentage kernel infection (P < 0.001). Fusarium and the Aspergillus spp. co-occurred in 30% of pearl millet samples.
Processed Samples
Contamination with Fusarium (15 of 21 samples; 71%) and the Aspergillus spp. (21 of 21 samples; 100%) was detected in the malts obtained from markets in Oshakati and Ondangwa (Table 3). The incidence of the Aspergillus spp. was significantly (P < 0.05) higher as compared to Fusarium spp. The highest contamination levels with the Aspergillus spp. were observed with NAM-11S (7 × 107 cfu/g), NAM-18M (1.1 × 107 cfu/g), NAM-19M (1.2 × 107 cfu/g) and NAM-25S (2 × 107 cfu/g) (P < 0.05). NAM-11S, NAM-18M and NAM-19M were obtained from the Oshakati market, whereas NAM-25S was obtained from the Ondangwa market. NAM-12S (6 × 105 cfu/g), NAM-19M (1.7 × 106 cfu/g), NAM-24S (3 × 105 cfu/g) and NAM-27S (range 6 × 105 cfu/g) exhibited the highest levels of Fusarium spp. contamination, with the highest incidence detected in pearl millet malt (P < 0.01). NAM-12S and NAM-19M were obtained from the Oshakati market, and NAM-24S and NAM-27S from the Ondangwa market. Fusarium and Aspergillus spp. co-occurred in 15 of 21 samples (74%) of malts sold at open markets.
Molecular Identification and Quantification of Mycotoxigenic Fusarium and Aflatoxigenic Aspergillus spp. in Sorghum and Pearl Millet Samples with qPCR
For optimization of the Fusarium qPCR reactions, the standard curves generated by applying F. verticillioides, F. proliferatum and F. graminearum, and species-specific primers showed linearity across the spectrum of the serial dilution concentrations used (Table S5). They also exhibited strong correlation coefficients, suggesting low inter-assay variability in all cases. The slope of the standard curves and the amplification efficiencies attained were within the acceptable range. To confirm the purity of the control sorghum and pearl millet samples, they were plated out onto potato PDA and analysed for fungal contamination and the results were negative in both respects. For optimization of the Aspergillus qPCR reactions the two standard curves created by the pairs FLAVIQ1/FLAQ2 and FLAVIQ1/PARQ2 lacked linearity across the range of concentrations used and displayed a correlation coefficient < 0.99 in all reactions. The slopes of the standard curves for A. flavus and A. parasiticus were − 2.365 and 0.039, respectively, corresponding to amplification efficiencies of 164% and 92,128 × 107%. The non-template control exhibited no amplification. All the values were out of range, and this served to confirm the lack of primer specificity. Aflatoxigenic Aspergillus spp. could therefore not be detected and quantified in samples by qPCR.
Fungal contamination was mainly observed in the malts and with F. verticillioides DNA (Table 4). Only a few (2 of 20 samples; 10%) raw whole grain samples contained F. verticillioides DNA opposed to 19 of 21 malts (90%). The highest incidence in sorghum malts was obtained with F. verticillioides (9 of 10 samples; 90%), followed by F. proliferatum (3 of 10 samples; 30%) and F. graminearum (2 of 10 samples; 20%) (P < 0.01). 56% of the contaminated sorghum malts originated from the Oshakati market and 44% from the Ondangwa market. In pearl millet malts, the highest contamination was obtained with F. verticillioides (10 of 11 samples; 91%) followed by F. graminearum (2 of 11 samples; 18%) (P < 0.01). No F. proliferatum was detected in pearl millet malts. 60% and 40% of the contaminated pearl millet malts originated from the Oshakati and Ondangwa markets, respectively. No correlation was observed between the incidence of Fusarium spp. as determined with morphological methods and that obtained with qPCR methods.
Detection and Quantification of Multiple Mycotoxins in Sorghum and Pearl Millet Samples with LC–MS/MS
The LC–MS/MS performance parameters for each mycotoxin are summarised in Table 2. The performance parameters for all mycotoxins were within acceptable ranges [39]. The results indicated that the control sorghum and pearl millet contained no mycotoxins. Selectivity of the method was confirmed by the absence of co-eluting peaks. The percentage recoveries for the individual mycotoxins (68–95%) in sorghum and pearl millet remained constant between runs. Results indicated that the extraction of FB was more effective (P < 0.05) from pearl millet than from sorghum. Replicate analysis of samples containing known amounts of the respective mycotoxins, resulted in means within 15% from the theoretical values, confirming the accuracy of the method. Coefficients of determination (R2), which indicates the degree of linearity of the respective mycotoxins’ calibration curves, were > 0.993. Mycotoxin concentrations in the FAPAS and Biopure certified quality control samples were within the ranges specified by the supplier for each mycotoxin during each LC–MS/MS run.
Contamination of Sorghum and Pearl Millet Samples with Multiple Mycotoxins
None of the analysed mycotoxins were detected in the raw whole grain samples collected postharvest, prior to storage and processing. These results correlate (R = 0.8–0.83) with the incidence of aflatoxigenic Aspergillus spp. (3 of 20 samples; 15%) detected with morphological methods (Table 3) and Fusarium spp. DNA determined with qPCR in raw whole grain samples, i.e. F. verticillioides (2 of 20 samples; 8%), F. proliferatum (0 of 20 samples) and F. graminearum (0 of 20 samples) (Table 4). With regards to the mycotoxin levels in processed samples, AFB1 was detected in sorghum malts (2 of 10 samples; 20%; 3–11 µg/kg) and pearl millet malts (6 of 11 samples; 55%; 4–14 µg/kg) (Table 5). These results correlated (R = 0.99) with the incidence of Aspergillus spp. in sorghum malts (10 of 10 samples; 100%) and pearl millet malts (10 of 10 samples, 100%) as determined with morphological methods (Table 3). FB was not detected in pearl millet malts (Table 5). FB1 (6 of 10 samples; 60%; 15–245 µg/kg) and FB2 (1 of 10 samples; 10%; 42 µg/kg) were present in the sorghum malts, which correlated (R = 0.88) with the incidence of F. verticillioides DNA in sorghum (9 of 10 samples; 90%) as determined with qPCR (Table 4). F. proliferatum DNA was detected in sorghum malts (3 of 10 samples; 30%), but not in pearl millet malts. One of the sorghum malts contained ZEA (3184 µg/kg) (Table 5). No detectable levels of DON were present in the malt samples. No F. graminearum DNA was detected in malt samples (Table 4).
Discussion
In Namibia, planting of sorghum and pearl millet crops takes place during November, and crops are harvested during June and July of the next year. After the growing season, crops are left to dry in the field [1, 42]. The stems are cut beneath the heads and the heads collected in harvesting baskets for further drying and threshing. As observed during the field study, threshing is performed close to the field, by hand, on hardened ground. Threshed heads are sun-dried on a threshing floor (Fig. S1) or on a raised wooden platform. Physical damage to heads exposes the powdery endosperm, thereby enhancing susceptibility to fungal infection during sun-drying, storage, and processing [1, 42]. Most households in the Oshana region store their sorghum and pearl millet for prolonged periods in traditional storage baskets (Fig. S2). The traditional granaries are large spherical woven baskets made of Mopani branches that are woven together using the bark. The internal surface is plastered using mud from ant and termite hills. The basket has a circular opening on top, which is closed by a lid and sealed by mud once loaded. Some storage baskets used in the North Central region of Namibia are made of Makalani palm leaves. Traditional methods of storage, which include the use of wood ash to guard from insect infection, could also be sources of fungal contamination [43]. Due to scarcity of trees in the Oshana region, some farmers have resorted to the use of plastic storage containers, which are commercially available.
A variety of traditional foods and beverages are prepared from sorghum. These include whole grain rice-type food, breads and pancakes, dumplings and couscous, porridges, gruels, opaque and cloudy beers, and non-alcoholic fermented beverages [1]. Throughout sub-Saharan Africa, sorghum is the grain of choice to produce traditional cloudy and opaque beers. The key ingredient of these beers is sorghum malt. Pearl millets are mainly used to prepare traditional fermented or unfermented porridges, and secondly for the brewing of traditional beers and wines [2]. Pearl millet-based gruels and steamed cakes are prepared for feeding infants and preschool children. Malted pearl millet in combination with legumes is used to prepare weaning foods. Malting is normally performed on household level and involves steeping in water (1–2 days), germination (4–5 days), and sun-drying [1, 22]. The temperature and moisture conditions during germination provide an ideal environment for fungi to proliferate, and could lead to an exponential increase in mycotoxin concentrations [22]. Milling of dried malts is traditionally performed by pounding with wooden pestles in a traditional mill until the grains are completely pulverized [22]. Some malts are transported and sold at open markets in urban areas or prepared for brewing at shebeens.
Limited information is available on the occurrence of mycotoxigenic fungi and mycotoxins along the complete sorghum and pearl millet production chain in Namibia. The present study addressed the occurrence of the main mycotoxin-producing fungi, i.e. mycotoxigenic Fusarium and aflatoxigenic Aspergillus spp. in the Oshana region of northern Namibia, mainly focussing on household grain and processed grain sold on open markets of Ondangwa and Oshakati. Samples were collected during a field study to Oshakati and Ondangwa during 2018. Traditional morphological methods as well as a validated qPCR method were used for the detection and quantification of Fusarium and Aspergillus spp. in the sorghum and pearl millet samples. A validated LC–MS/MS method was used to determine the concentrations of multiple mycotoxins in samples.
No correlation existed between the incidence of Fusarium spp. in samples obtained with morphological methods and that determined with qPCR. Most members of the Fusarium genus are morphologically similar or are cryptic species [44]. This makes it increasingly challenging and inaccurate to rely only on morphological features for identification [26, 44]. Traditional methods for identification and characterization of mycotoxigenic fungi are currently complemented with molecular based approaches such as PCR [45]. PCR-based genotyping based on sequence variability and the presence of certain genes such as EF1α, translation elongation factor 1-alpha (TEF1-α), IGS, and mycotoxin biosynthetic genes such as FUM1, TRI13 and TRI17 has become useful and more reliable fungal identification methods [46]. In this study, the use of species-specific primers allowed the detection and quantification of mycotoxigenic Fusarium spp. in samples. It should, however, be noted that qPCR can only be applied to grain samples to detect and quantify fungal species (i) that are known, (ii) for which species-specific primers are available, and (iii) for which species-specific primers have been optimized for the relevant grain matrix. It should therefore be used to complement mycological methods, to ensure that the correct fungal species are targeted.
High quality DNA was extracted from sorghum and pearl millet samples as well as from liquid cultures of reference Fusarium and Aspergillus spp. The DNA was extracted from 2 g of each sample. This was done to reduce the errors caused by non-uniform distribution of Fusarium spp. in samples. Although a higher amount of plant tissue might be desirable, it could saturate the mini extraction column and have a negative effect on the efficacy of DNA extractions. The DNA reading on the Thermo Scientific Nanodrop 2000 Spectrophotometer showed all the DNA samples used in these experiments have high molecular weight fragments with an A260/280 ratio between 1.8 and 2.0. To ensure that both the fungal and plant cell walls were properly broken and a high yield of gDNA was obtained during extraction, a liquid nitrogen homogenize step was included in the protocol for both Fusarium and Aspergillus spp. strains as well as for the sorghum and pearl millet samples. Phenolics are considered as the main contaminants in plant and fungal DNA preparation [47]. Phenolics, being strong oxidizing agents, decrease the yield and purity of DNA by binding covalently to the isolated DNA, thus inhibiting further enzymatic reactions of DNA, such as PCRs. The CTAB and the addition of PVP, an antioxidant, to the extraction buffer assist in eliminating phenolics in DNA extracted from plants and fungi. To ensure complete removal of phenols from the fungal gDNA, PCI and CI steps were added as modifications to the protocol provided by the manufacturer. Unfortunately, the Aspergillus spp. could not be detected in the sorghum and pearl millet samples, due to primer non-specificity. Aflatoxigenic Aspergillus spp. could, however, successfully be detected and quantified with morphological methods using Aspergillus differentiation agar.
No mycotoxins were detected in any of the raw sorghum and pearl millet whole grain samples collected from 10 households of smallholder farmers in Oshakati, postharvest, prior to storage and processing. Contrary to the raw whole grain samples, the processed samples contained mycotoxins (AFB1, FB and ZEA) of which MLs in food have been set for many countries by the Codex Alimentarius Commission [24, 48] and the European Commission (EC) [49]. Two of 10 (20%) sorghum and 6 of 11 (55%) pearl millet malts contained AFB1 above the ML of 2 µg/kg for processed cereals set by the EC [49]. A high correlation was observed between the incidences of Aspergillus spp. and AFB1 contamination. Comparable levels of AFB1 and FB1 were previously detected in sorghum malts used for the brewing of oshikundu [3], amaludo [22] and otombo in Namibia. AFB1 (4.5 ± 5.5 µg/kg) and FB1 (28.2 ± 33.3 μg/kg) were detected in sorghum malts used for brewing of oshikundu [3]. AFB1 (2.87–15.1 µg/kg) and FB1 (29.12–61.4 µg/kg) were detected in sorghum malts used for brewing omalodu and otombo [22]. In the current study, one of the sorghum malts contained ZEA above the MLs set by the EC [49] for cereal-based food and baby foods for infants and young children. Contamination by ZEA often co-occurs with DON [50]. However, there were non-detectable levels of DON contamination in the processed samples. The results of this study indicated that contamination of the processed samples occurred postharvest, possibly during storage and/or transportation and processing, due to an environment that favoured the proliferation of certain fungal species and subsequent production of mycotoxins.
Co-contamination of food and co-exposure of particularly young children to multiple mycotoxins in their staple diet have been extensively reported in certain communities in African and Latin American countries [5, 51, 52]. The Joint Food and Agricultural Organisation of the United Nations (FAO)/World Health Organisation (WHO) Expert Committee on Food Additives (JECFA) has expressed concern about the possible interaction between AFB1, a human genotoxin and carcinogen, and FBs, which have the potential to induce regenerative cell proliferation [53]. JECFA has determined Provisional Maximum Tolerable Daily Intake (PMTDI) levels for total FB of 2 µg/kg body weight (bw) per day [54]. It has, however, been demonstrated that the day-to-day high consumption levels of grains containing low concentrations of FB could result in PMTDI levels above the threshold of 2 µg/kg bw per day [55]. High consumption levels and chronic exposure to contaminated grains may enhance the negative health effects, especially in immunocompromised individuals [56].
Conclusions
Raw whole grain sorghum and pearl millet produced by smallholder farmers in northern Namibia contained none of the mycotoxins analysed. Aflatoxigenic Aspergillus spp. were, however, detected in all sorghum and pearl millet malts. F. verticillioides was the predominant Fusarium sp. present in malts. Eight of 21 (38%) of malts contained AFB1 above the ML set by the EC [49]. The co-occurrence of Fusarium and Aspergillus spp. in processed sorghum and pearl millet as well as chronic exposure of the communities to AFB1 and FB in their staple diet is a serious concern. The malts are sold at markets and used to prepare a variety of food and beverages, including weaning foods.
Risk management involving the implementation of control methods to reduce the levels of mycotoxins in sorghum and pearl millet malts is essential. By critically monitoring the grain production process from planting, through to harvesting, processing, transportation, and marketing, the sources of contamination and critical control points could be identified and managed. Good agricultural management, both pre-harvest and postharvest, and the implementation of hazard analysis critical control point (HACCP) systems will assist in reducing fungal growth and mycotoxin contamination [57]. Several technological methods have been developed to manage pre- and postharvest fungal growth and mycotoxin production, i.e., methods involving clay minerals, plant extracts, antioxidants, biocontrol microorganisms and enzymes [58]. In Africa, there are limited resources and a scarcity of sophisticated technologies. The WHO [59] made recommendations for reduction of mycotoxins in staple grains applicable to rural subsistence farming communities. Community-based practical and integrated interventions are relevant and need to be implemented. This could involve peer-to-peer training to improve awareness and knowledge, dissemination of community-specific good agricultural practices, hand sorting, crushing, dehulling, washing, winnowing, and milling of grains [58], as well as hermitic storage practices [60].
This study presented a risk assessment of the contamination by mycotoxigenic fungi and mycotoxins of the staple grains, sorghum and pearl millet, consumed by smallholder farming communities in the Oshana region of northern Namibia. The results indicated that mycotoxigenic Fusarium and aflatoxigenic Aspergillus spp. colonize mainly processed sorghum and pearl millet with the occurrence of the carcinogenic aflatoxin and fumonisin mycotoxins. To determine the full extent of contamination of staple grains with multiple mycotoxins in Namibia, surveillance studies should be extended to more regions. These studies should be followed up with characterization of the dietary exposure of vulnerable populations by considering the consumption levels of the grains and corresponding contamination levels with multiple mycotoxins [61]. Understanding the impact of mycotoxins on human health is critical to further improve the risk management processes through monitoring, management, the development of informed policy strategies and eventually the implementation of regulations. This could ultimately contribute to food safety and security in northern Namibia where communities are exposed to multiple mycotoxins in their staple diet.
Data Availability
Available on request.
Code Availability
Not applicable.
References
Taylor JR (2003) Overview: importance of sorghum in Africa. In AFRIPRO workshop on the proteins of sorghum and millets: enhancing nutritional and functional properties for Africa. Pretoria, South Africa 2–4 April 2003. http://afripro.org.uk/papers/Paper01Taylor.pdf. Accessed 15 Sep 2021
Laminu HH, Modu S, Numan AI (2011) Production, in vitro protein digestibility, phytate content and acceptability of weaning foods prepared from pearl millet (Pennisetum typhoideum) and cowpea (Vigna unguiculata). Int J Nutr Metab 3:109–113
Misihairabgwi JM, Ishola A, Quaye I, Sulyok M, Krska R (2018) Diversity and fate of fungal metabolites during the preparation of oshikundu, a Namibian traditional fermented beverage. World Mycotoxin J 11:471–481
Alberts J, Rheeder J, Gelderblom W, Shephard G (2019) Rural subsistence maize farming in South Africa: risk assessment and intervention models for reduction of exposure to fumonisin mycotoxins. Toxins 11(6):334. https://doi.org/10.3390/toxins11060334
Shephard GS, Burger HM, Rheeder JP, Alberts JF, Gelderblom WCA (2019) The effectiveness of regulatory maximum levels for fumonisin mycotoxins in commercial and subsistence maize crops in South Africa. Food Control 97:77–80
Marasas WFO (1995) Fumonisins: their implications for human and animal health. Nat Toxins 3:193–198
Rushing BR, Selim MI (2019) Aflatoxin B1: a review on metabolism, toxicity, occurrence in food, occupational exposure, and detoxification methods. Food Chem Toxicol 124:81–100
International Agency for Research on Cancer (IARC) (2012) Improving public health through mycotoxin control. In: Pitt I, Wild CP, Baan R, Gelderblom WCA, Miller JD, Riley RT, Wu F (eds) World Health Organisation, Geneva. http://scabusa.org/pdfs/IARC_STP-158_Dec-2012.pdf Accessed 20 Aug 2021
Pitt JI (2012) Improving public health through flavorful eating. Nutr Today 49:S2–S3
Smith LE, Stolzfus RJ, Prendergast A (2012) Food chain mycotoxin exposure, gut health, and impaired growth: a conceptual framework. Adv Nutr 3:526–531
Riedel S, Abel S, Swanevelder S, Gelderblom WCA (2015) Induction of an altered lipid phenotype by two cancer promoting treatments in rat liver. Food Chem Toxicol 78:96–104
Pinto ACSM, De Pierri CR, Evangelista AG, Gomes ASDLPB, Luciano FB (2022) Deoxynivalenol: toxicology, degradation by bacteria, and phylogenetic analysis. Toxins 14(2):90. https://doi.org/10.3390/toxins14020090
Zhang G-L, Feng Y-L, Song J-L, Zhou X-S (2018) Zearalenone: a mycotoxin with different toxic effect in domestic and laboratory animals’ granulosa cells. Front Genet. https://doi.org/10.3389/fgene.2018.00667
Lombard MJ (2014) Mycotoxin exposure and infant and young child growth in Africa: what do we know? Ann Nutr Metab 64:442–452
Vismer HF, Shephard GS, Rheeder JP, van der Westhuizen L, Bandyopadhyay R (2015) Relative severity of fumonisin contamination of cereal crops in West Africa. Food Addit Contam 32:1952–1958
Ssepuuya G, Van Poucke C, Ediage EN, Mulholland C, Tritscher A, Verger P, Kenny M, Bessy C, De Saeger S (2018) Mycotoxin contamination of sorghum and its contribution to human dietary exposure in four sub-Saharan countries. Food Addit Contam 35:1384–1393
Janse van Rensburg B, McLaren NW, Viljoen A, Flett BC (2011) Aflatoxin and fumonisin on sorghum grain from commercial production areas of South Africa. S Afr J Plant Soil 28:236–238
Beukes I, Rose LJ, Shephard GS, Flett BC, Viljoen A (2017) Mycotoxigenic Fusarium species associated with grain crops in South Africa: a review. S Afr J Sci 113:3–4. https://doi.org/10.17159/sajs.2017/20160121
Misihairabgwi JM, Ezekiel CN, Sulyok M, Shephard GS, Krska R (2019) Mycotoxin contamination of foods in Southern Africa: a 10-year review (2007–2016). Crit Rev Food Sci Nutr 59:43–58
Klaasen JA, Nelson PE (1997) Identity of Fusarium nygamai isolates with long and short microconidial chains from millet, sorghum and soil in Africa. Mycopathologia 140:171–176
Nkwe DO, Taylor JE, Siame BA (2005) Fungi, aflatoxins, fumonisin B1 and zearalenone contaminating sorghum-based traditional malt, wort and beer in Botswana. Mycopathologia 160:177–186
Nafuka SN, Misihairabgwi JM, Bock R, Ishola A, Sulyok M, Krska R (2019) Variation of fungal metabolites in sorghum malts used to prepare Namibian traditional fermented beverages Omalodu and Otombo. Toxins 11:165. https://doi.org/10.3390/toxins11030165
Shephard GS, Marasas WFO, Burger H-M, Somdyala NIM, Rheeder JP, Van der Westhuizen L, Gatyeni P, Van Schalkwyk DJ (2007) Exposure assessment for fumonisins in the former Transkei region of South Africa. Food Addit Contam 24:621–629
Food and Agricultural Organization of the United Nations (FAO) (2003) Worldwide regulations of mycotoxins in food and feed in 2003. https://www.fao.org/3/y5499e/y5499e00.pdf. Accessed 4 Jul 2021
Van Egmund HP, Jonker MA (2005) Worldwide regulations for mycotoxins in food and feed in 2003. Summary of study, carried out for the Food and Agriculture Organization (FAO). Food and Nutrition Paper No. 81. Food and Agricultural Organisation. https://www.rivm.nl/bibliotheek/digitaaldepot/23661ADC.pdf. Accessed 15 Aug 2021
Leslie JF, Summerell BA (2006) The Fusarium laboratory manual. Wiley-Blackwell, Ames
Safrinet (1999) Collecting and preserving fungi. http://www.mires-and-peat.net/map07/map_07_11.pdf. Accessed 24 Jan 2021
Rheeder JP, Van der Westhuizen L, Imrie G, Shephard GS (2016) Fusarium species and fumonisins in subsistence maize in the former Transkei region, South Africa: a multi-year study in rural villages. Food Addit Contam 9:176–184
Burgess LW, Summerell BA, Bullock S, Gott KP, Backhouse D (1994) Laboratory manual for Fusarium research, 3rd edn. University of Sydney/Royal Botanic Gardens, Sydney
Nirenberg HI, O’Donnell K (1998) New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia 90:434–458
Pitt JI, Hocking AD, Glenn DR (1983) An improved medium for the detection of Aspergillus flavus and A. parasiticus. J Appl Bacteriol 54:109–114
Van Wyk PS, Scholtz DI, Los O (1986) A selective medium for the isolation of Fusarium spp. from soil debris. Phytophylactica 18:67–69
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Nicolaisen M, Suproniene S, Nielsen LK, Lazzaro I, Spliid NH, Justesen AF (2009) Real-time PCR for quantification of eleven individual Fusarium species in cereals. J Microbiol Methods 76:234–240
Sardiñas N, Vázquez C, Gil-Serna J, González-Jaén MT, Patiño B (2010) Specific detection of Aspergillus parasiticus in wheat flour using a highly sensitive PCR assay. Food Addit Contam 27:853–858
Boutigny AL, Beukes I, Small I, Zühlke S, Spiteller M, Van Rensburg BJ, Flett B, Viljoen A (2011) Quantitative detection of Fusarium pathogens and their mycotoxins in South African maize. Plant Pathol 61:522–531
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Alberts J, Schatzmayr G, Moll W-D, Davids I, Rheeder J, Burger H-M, Shephard G, Gelderblom W (2019) Detoxification of the fumonisin mycotoxins in maize: an enzymatic approach. Toxins 11:523. https://doi.org/10.3390/toxins11090523
United States Department of Health and Human Services, Food and Drug Administration (FDA), USA (2018) Guidance for Industry. Bioanalytical Method Validation. https://www.fda.gov/files/drugs/published/Bioanalytical-Method-Validation-Guidance-for-Industry.pdf. Accessed 10 Aug 2022
NCSS (2019) Statistical software (2019) NCSS, LLC. Kaysville. https://www.ncss.com/software/ncss/. Accessed 30 May 2021
Harris R (2018) An introduction to R. Quant Geogr. https://doi.org/10.4135/9781473920446
Mallet M, du Plessis P (2001) Mahangu post-harvest systems: a summary of current knowledge about pearl millet post-harvest issues in Namibia: Research report. Ministry of Agriculture, Water, and Rural Development, Directorate of Planning and Namibian Agronomic Board. http://the-eis.com/elibrary/sites/default/files/downloads/literature/Mahangu_Post-Harvest_Systems.pdf. Accessed 30 Aug 2021
Tangni EK, Larondelle Y (2002) Malts, moulds and mycotoxins. In: Bacteria, Yeasts and Moulds in Malting and Brewing: Proceedings of the Xth Symposium “Chair J. de Clerck”, 15–18 September 2002
Mavhunga M (2013) Fusarium graminearum mycotoxins associated with grain mould of maize and sorghum in South Africa. PhD thesis, University of the Free State, Bloemfontein
McClenny N (2005) Laboratory detection and identification of Aspergillus species by microscopic observation and culture: the traditional approach. Med Mycol 43(1):125–128. https://doi.org/10.1080/13693780500052222
Sampietro DA, Marín P, Iglesias J, Presello DA, Vattuone MA, Catalan CAN, Gonzalez Jaen MT (2010) A molecular based strategy for rapid diagnosis of toxigenic Fusarium species associated to cereal grains from Argentina. Fungal Biol 114:74–81
Calderón-Cortés N, Quesada M, Cano-Camacho H, Zavala-Páramo G (2010) A simple and rapid method for DNA isolation from xylophagous insects. Int J Mol Sci 11:5056–5064
Codex Alimentarius Commission (2019) General standard CXS 193–1995 for contaminants and toxins in food and feed. Food and Agriculture Organization of the United Nations/World Health Organization. https://www.fao.org/fao-who-codexalimentarius/sh-proxy/en/?lnk=1&url=https%253A%252F%252Fworkspace.fao.org%252Fsites%252Fcodex%252FStandards%252FCXS%2B193-1995%252FCXS_193e.pdf. Accessed 15 Jan 2022
European Commission (EC) (2006) Commission regulation (EC) No 1881/2006 of 19 December 2006 setting maximum levels for certain contaminants in foodstuffs. Off J Eur Union L364:5–24
Ferrigo D, Raiola A, Causin R (2016) Fusarium toxins in cereals: Occurrence, legislation, factors promoting the appearance and their management. Molecules 21:627. https://doi.org/10.3390/molecules21050627
Ezekiel CN, Warth B, Ogara IM, Abia WA, Ezekiel VC, Atehnkeng J, Sulyok M, Turner PC, Tayo GO, Krska R, Bandyopadhyay R (2014) Mycotoxin exposure in rural residents in northern Nigeria: A pilot study using multi-urinary biomarkers. Environ Int 66:138–145
Torres O, Matute J, Gelineau-Van Waes J, Maddox JR, Gregory SG, Ashley-Koch AE, Showker JL, Voss KA, Riley RT (2015) Human health implications from co-exposure to aflatoxins and fumonisins in maize-based foods in Latin America: Guatemala as a case study. World Mycotoxin J 8:143–159
World Health Organization (WHO) (2017) Fumonisins. Evaluation of certain contaminants in food. WHO technical report series 1002, Prepared by the 83rd meeting of the Joint FAO/WHO Expert Committee on Food Additives (JECFA). WHO, Geneva, pp 55–73
Joint FAO/WHO Expert Committee on Food Additives (JECFA) (2012) Fumonisins. In: Joint FAO/WHO Expert Committee on food additives (ed) Safety evaluation of certain food additives and contaminants. ISBN 9789241660655
Burger H-M, Lombard MJ, Shephard GS, Danster-Christians N, Gelderblom WCA (2014) Development and evaluation of a sensitive mycotoxin risk assessment model (MYCORAM). Tox Sci 141:387–397
Alberts JF, Lilly M, Rheeder JP, Burger H-M, Shephard GS, Gelderblom WCA (2017) Technological and community-based methods to reduce mycotoxin exposure. Food Control 73:101–109
Food and Agriculture Organization (FAO) of the United Nations (1997) Hazard analysis and critical control point (HACCP) system and guidelines for its application. Codex Alimentarius Commission. https://www.fao.org/3/y1579e/y1579e03.htm Accessed 15 Jan 2022
Alberts JF, van Zyl WH, Gelderblom WCA (2016) Biologically based methods for control of fumonisin-producing Fusarium species and reduction of the fumonisins. Front Microbiol 7:548. https://doi.org/10.3389/fmicb.2016.00548
World Health Organization (WHO) (2009) Environmental health criteria 240, principles and methods for the risk assessment of chemicals in food. FAO, WHO, Geneva
Ngwenyama P, Myumi BM, Nyanga LK, Stathers TE, Siziba S (2020) Comparative performance of five hermetic bag brands during on-farm smallholder cowpea (Vigna unguiculata L. Walp) storage. J Stored Prod Res 88:1–10. https://doi.org/10.1016/j.jspr.2020.101658
Tshalibe RS, Rheeder JP, Alberts JF, Taljaard-Krugell C, Gelderblom WCA, Shephard GS, Lombard MJ, Burger H-M (2020) Multi-mycotoxin exposure of children (0–24 months) in rural maize-subsistence farming areas of the Eastern Cape Province, South Africa. World Mycotoxin J 3:401–410
Acknowledgements
The authors thank Ms G Imrie and Mr T Leukes for laboratory assistance.
Funding
Open access funding provided by Cape Peninsula University of Technology. Cape Peninsula University of Technology (South Africa) University Research Funding (Ref. 2017/2018 URF).
Author information
Authors and Affiliations
Contributions
Conceptualization: JFA, JPR, JMM; Formal analysis and investigation: JFA, CRK, ML, JPR, JMM; Funding acquisition: JPR; Methodology: JFA, ML, JPR; Project administration: JFA; Resources: JFA, JPR, JMM; Supervision: JFA, ML, JPR; Validation: JFA, CRK, ML; Writing original draft: JFA, CRK; Writing, review and editing: JFA, CRK, ML, JMM.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
Ethical clearance for the study was obtained from the Health and Wellness Research Ethics Committee of the Cape Peninsula University of Technology, South Africa (Approval Registration No. CPUT/HW-REC 2018/H5).
Consent to Participate
Smallholder farmer participants signed informed consent forms for voluntary permission for samples to be taken from their households and for the protection of their household privacy.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kaela, C.R., Lilly, M., Rheeder, J.P. et al. Mycological and Multiple Mycotoxin Surveillance of Sorghum and Pearl Millet Produced by Smallholder Farmers in Namibia. Curr Microbiol 80, 164 (2023). https://doi.org/10.1007/s00284-023-03263-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03263-7