Abstract
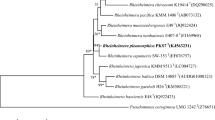
Two Gram stain-negative bacterial strains designated Jin1T and Jin2T were isolated from artificial pond water in the Republic of Korea and investigated in the present study using a polyphasic taxonomic approach. Phylogenetic analysis of the 16S rRNA gene sequences indicated that these two strains belonged to the genera Chryseolinea and Piscinibacter due to their highest similarities with the 16S rRNA gene sequences of Chryseolinea serpens RYGT (98.7%) and Piscinibacter aquaticus IMCC1728T (97.1%), respectively. The dDDH and ANI values were 18.1–20.5% and 68.9–76.8% between whole-genome sequences of strain Jin1T and type trains of the selected taxa, Chryseolinea species, and 20.1–20.2% and 75.5–76.1% between those of strain Jin2T and the type strains of the selected taxa, Piscinibacter species, respectively. A threshold AAI value of 44.7–64.8% for the species boundary (95–96%) was established for strains Jin1T, Jin2T, and type strains of other species involved in the system incidence, which confirms that strains Jin1T and Jin2T represent two new species of the genera Chryseolinea and Piscinibacter, respectively. Based on the phylogenetic, chemotaxonomic, and phenotypic analyses, strains Jin1T and Jin2T represent novel species of the genera Chryseolinea and Piscinibacter, respectively, for which the names Chryseolinea lacunae sp. nov. (type strain Jin1T = KCTC 82562T = NBRC 114837T) and Piscinibacter lacus sp. nov. (type strain Jin2T = KCTC 82556T = NBRC 114838T) have been proposed.



Similar content being viewed by others
References
Kim JJ, Alkawally M, Brady AL et al (2013) Chryseolinea serpens gen. nov., sp. nov., a member of the phylum Bacteroidetes isolated from soil. Int J Syst Evol Microbiol 63:654–660. https://doi.org/10.1099/IJS.0.039404-0
Parte AC (2018) LPSN - list of prokaryotic names with standing in nomenclature (Bacterio.net), 20 years on. Int J Syst Evol Microbiol 68:1825–1829
Wang J, jing, Chen Q, Li Y zhong, (2018) Chryseolinea flava sp. nov., a new species of Chryseolinea isolated from soil. Int J Syst Evol Microbiol 68:3518–3522. https://doi.org/10.1099/IJSEM.0.003022
Song J, Cho JC (2007) Methylibium aquaticum sp. nov., a betaproteobacterium isolated from eutrophic freshwater pond. Int J Syst Evol Microbiol 57:2125–2128. https://doi.org/10.1099/IJS.0.65179-0
Stackebrandt E, Verbarg S, Frühling A et al (2009) Dissection of the genus Methylibium: reclassification of Methylibium fulvum as Rhizobacter fulvus comb. nov., Methylibium aquaticum as Piscinibacter aquaticus gen. nov., comb. nov. and Methylibium subsaxonicum as Rivibacter subsaxonicus gen. nov., comb. nov. and emended descriptions of the genera Rhizobacter and Methylibium. Int J Syst Evol Microbiol 59:2552–2560. https://doi.org/10.1099/IJS.0.008383-0
Chen DZ, Yu NN, Chu QY et al (2018) Piscinibacter caeni sp. nov., isolated from activated sludge. Int J Syst Evol Microbiol 68:2627–2632. https://doi.org/10.1099/IJSEM.0.002891
Cho SH, Lee HJ, Jeon CO (2016) Piscinibacter defluvii sp. nov., isolated from a sewage treatment plant, and emended description of the genus Piscinibacter Stackebrandt et al. 2009. Int J Syst Evol Microbiol. 66:4839–4843. https://doi.org/10.1099/IJSEM.0.001438
Kim I, Chhetri G, Kim J et al (2020) Lewinella aurantiaca sp. nov., a carotenoid pigment-producing bacterium isolated from surface seawater. Int J Syst Evol Microbiol 70:6180–6187. https://doi.org/10.1099/ijsem.0.004515
Kim I, Chhetri G, Kim J et al (2020) Reinekea thalattae sp. nov., a New species of the genus Reinekea isolated from surface seawater in Sehwa beach. Curr Microbiol 7712(77):4174–4179. https://doi.org/10.1007/S00284-020-02224-8
Yoon SH, Ha SM, Kwon S et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/IJSEM.0.001755
Altschul SF, Madden TL, Schäffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/NAR/25.17.3389
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/MOLBEV/MSY096
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 162(16):111–120. https://doi.org/10.1007/BF01731581
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol 20:406–416. https://doi.org/10.1093/sysbio/20.4.406
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Bankevich A, Nurk S, Antipov D et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Parks DH, Imelfort M, Skennerton CT et al (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Research 25:1043–1055. https://doi.org/10.1101/gr.186072.114
Chun J, Oren A, Ventosa A et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466. https://doi.org/10.1099/ijsem.0.002516
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 141(14):1–14. https://doi.org/10.1186/1471-2105-14-60
Blin K, Shaw S, Steinke K et al (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. https://doi.org/10.1093/NAR/GKZ310
Huerta-Cepas J, Szklarczyk D, Heller D et al (2019) eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. https://doi.org/10.1093/NAR/GKY1085
Buck JD (1982) Nonstaining (KOH) method for determination of gram reactions of marine bacteria. Appl Environ Microbiol 44:992–993. https://doi.org/10.1128/aem.44.4.992-993.1982
Bernardet J-F, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070. https://doi.org/10.1099/00207713-52-3-1049
Chhetri G, Kim J, Kim I et al (2021) Flavobacterium baculatum sp. nov., a carotenoid and flexirubin-type pigment producing species isolated from flooded paddy field. Int J Syst Evol Microbiol 71:004736. https://doi.org/10.1099/IJSEM.0.004736
Hiraishi A, Ueda Y, Ishihara J, Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–459. https://doi.org/10.2323/jgam.42.457
Collins MD, Jones D (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol Rev 45:316–354
Komagata K, Suzuki KI (1988) 4 Lipid and cell-wall analysis in bacterial systematics. Methods in Microbiology, vol 19. Elsevier, Amsterdam, pp 161–207
Fautz E, Reichenbach H (1980) A simple test for flexirubin-type pigments. FEMS Microbiol Lett 8:87–91. https://doi.org/10.1111/j.1574-6968.1980.tb05056.x
Kim I, Chhetri G, Kim J et al (2021) Nocardioides donggukensis sp. nov. and Hyunsoonleella aquatilis sp. nov., isolated from Jeongbang Waterfall on Jeju Island. Int J Syst Evol Microbiol. 71:005176. https://doi.org/10.1099/IJSEM.0.005176
Lee SA, Kim Y, Sang MK, Song J, Know SW, Weon HY (2019) Chryseolinea soli sp. nov., isolated from soil. J Microbiol. 57(2):122–126. https://doi.org/10.1007/S12275-019-8562-4
Acknowledgements
This work was supported by a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR202002203) and by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2022R1F1A1070108).
Funding
This work was supported by a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR202002203) and by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2022R1F1A1070108).
Author information
Authors and Affiliations
Contributions
IK, designed the study, isolated the bacterium, performed the phenotypic and biochemical experiments, analyzed the data, and wrote the original draft; GC, JK, and YS helped with the analysis of taxonomic data; TS designed and supervised the study and edited the original draft; TS designed and supervised the study and edited the original draft.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical Approval
This study does not describe any experimental work related to humans.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kim, I., Chhetri, G., Kim, J. et al. Chryseolinea lacunae sp. nov. and Piscinibacter lacus sp. nov. Isolated from Artificial Pond Water. Curr Microbiol 80, 25 (2023). https://doi.org/10.1007/s00284-022-03133-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03133-8