Abstract
l-Arabinofuranosides with β-linkages are present in several plant molecules, such as arabinogalactan proteins (AGPs), extensin, arabinan, and rhamnogalacturonan-II. We previously characterized a β-l-arabinofuranosidase from Bifidobacterium longum subsp. longum JCM 1217, Bll1HypBA1, which was found to belong to the glycoside hydrolase (GH) family 127. This strain encodes two GH127 genes and two GH146 genes. In the present study, we characterized a GH146 β-l-arabinofuranosidase, Bll3HypBA1 (BLLJ_1848), which was found to constitute a gene cluster with AGP-degrading enzymes. This recombinant enzyme degraded AGPs and arabinan, which contain Araf-β1,3-Araf structures. In addition, the recombinant enzyme hydrolyzed oligosaccharides containing Araf-β1,3-Araf structures but not those containing Araf-β1,2-Araf and Araf-β1,5-Araf structures. The crystal structures of Bll3HypBA1 were determined at resolutions up to 1.7 Å. The monomeric structure of Bll3HypBA1 comprised a catalytic (α/α)6 barrel and two β-sandwich domains. A hairpin structure with two β-strands was observed in Bll3HypBA1, to extend from a β-sandwich domain and partially cover the active site. The active site contains a Zn2+ ion coordinated by Cys3-Glu and exhibits structural conservation of the GH127 cysteine glycosidase Bll1HypBA1. This is the first study to report on a β1,3-specific β-l-arabinofuranosidase.
Key points
• β1,3-l-Arabinofuranose residues are present in arabinogalactan proteins and arabinans as a terminal sugar.
• β-l-Arabinofuranosidases are widely present in intestinal bacteria.
• Bll3HypBA1 is the first enzyme characterized as a β1,3-linkage-specific β-l-arabinofuranosidase.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
l-Arabinose occurs in four distinct forms in plant polysaccharides: α- and β-l-arabinofuranose (Araf) and l-arabinopyranose (Arap). Although β-Araf occurs in significantly less amount than α-Araf in plant carbohydrate polymers, it is present as β-l-arabinooligosaccharide (β-AOS) chains in extensin (e.g., Araf-β1,2-Araf-β1,2-Araf-β1-hydroxyproline (Hyp); Ara3-Hyp). β-Araf is also present as a terminal sugar in plant polysaccharides, e.g., terminal β1,2- and β1,5-Araf are present in the complicated sugar chain of rhamnogalacturonan-II (RG-II) in pectin (Pellerin et al. 1996). Moreover, terminal Araf-β1,3-Araf-α1 structures are found in several plant polysaccharides, including arabinan from sugar beet (Wefers et al. 2017) and quinoa seeds (Wefers et al. 2014), arabinoxylan from green leaves of Lauraceae (Das et al. 2013), arabinoxyloglucan from tomato cultured cells (York et al. 1996), and arabinogalactan proteins (AGPs) from rice anthers (Kawaguchi et al. 1996). β-Arap is also present as a terminal sugar in plant polysaccharides, including type-II arabinogalactan (AG) from larch (Ponder and Richards 1997), AGPs from wheat (Tryfona et al. 2010), and gum arabic (Tischer et al. 2002). α-Arap is found only as a constituent sugar of RG-II in plant polysaccharides (Pellerin et al. 1996).
Pfam DUF1680 (PF07944), which was renamed to Glyco_hydro_127, contains 24,000 proteins that are distributed among 7482 bacterial, fungal, and plant species. HypBA1 (Bll1HypBA1; encoded by BLLJ_0211) is the first characterized GH127 β-l-arabinofuranosidase (EC 3.2.1.185) (Fujita et al. 2014b). Bll1HypBA1 from Bifidobacterium longum subsp. longum JCM 1217 degrades Araf-β1,2-Ara released from Ara3-Hyp by GH121 β-l-arabinobiosidase (HypBA2: BLLJ_0212) (Fujita et al. 2011). Crystallographic studies of Bll1HypBA1 have revealed that a cysteine residue serves as the catalytic nucleophile (Ishiwata et al. 2022b; Ito et al. 2014; Maruyama et al. 2022; McGregor et al. 2021). In addition to GH137 and GH142 β-l-arabinofuranosidases for RG-II degradation in Bacteroides thetaiotaomicron (Ndeh et al. 2017), GH146 was established after characterization of BT0349 as an β-l-arabinofuranosidase for arabinan-derived arabinotetraose and β1,2-l-arabinobiose (Luis et al. 2018). We also characterized a GH146 member, XCV2724, from Xanthomonas euvesicatoria (XeHypBA1) (Nakamura et al. 2018). This enzyme hydrolyzed Araf-β1-Hyp predominately and partially for Araf-β1,2-Ara (Ishiwata et al. 2023). In addition to two GH127 members (Bll1HypBA1 and BLLJ_1826), B. longum subsp. longum JCM 1217 encodes two GH146 members (BLLJ_1848 and BLLJ_0089). BLLJ_0089 was recently characterized as a β-l-arabinofuranosidase (Bll4HypBA1) for Araf-β1-linked Hyp on the backbone of Hyp-rich glycoprotein (HRGP) (Ishiwata et al. 2023). At present, the three-dimensional structure of only one enzyme (BT0349) in the GH146 family has been reported (Luis et al. 2018; McGregor et al. 2021). In the present study, we characterized the recombinant BLLJ_1848 as a β-1,3-specific GH146 β-l-arabinofuranosidase (Bll3HypBA1) using natural and synthetic substrates. In addition, we performed X-ray crystallography of Bll3HypBA1.
Materials and methods
Substrates
Larch wood AG was purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). Gum arabic from Acacia senegal was purchased from Sigma-Aldrich (St. Louis, MO, USA). p-Nitrophenyl (pNP) β-l-arabinofuranoside was synthesized as described in a previous study (Kaeothip et al. 2013a). Young rice panicles, cultivar Nikomaru, were harvested from experimental farms at the Faculty of Agriculture, Kagoshima University (Kagoshima, Japan). According to a previous study (Kawaguchi et al. 1996), l-Araf-β1,3-l-Araf-α1,3-Gal-β1,6-Gal (Araf-β1,3-ArafGal2) and AGP from rice anthers were prepared from young rice panicles. l-Araf-α1,5-[l-Araf-β1,3-l-Araf-α1,3-]-l-Araf-α1,5-l-Ara (Araf-β1,3-Araf4) from quinoa seed arabinan was prepared through endo-1,5-α-l-arabinanase treatment as described by Wefers et al. (Wefers et al. 2014). These oligosaccharides were further purified using HPLC on a Cosmosil PBr column (Nacalai Tesque Inc., Kyoto, Japan) as described previously (Sasaki et al. 2021). The fractions containing oligosaccharides were collected and analyzed using MALDI-TOF MS (Bruker Daltonics, Leipzig, Germany).
l-Araf-β1,2-l-Araf-α1-OMe (Araf-β1,2-Araf-α-OMe), l-Araf-β1,3-l-Araf-α1-OMe (Araf-β1,3-Araf-α-OMe), and l-Araf-β1,5-l-Araf-α1-OMe (Araf-β1,5-Araf-α-OMe) were previously prepared stereospecifically (Ishiwata et al. 2022a) via 1,2-cis selective β-arabinofuranosylation using 2-naphthylmethyl ether-mediated intramolecular aglycon delivery (Ishiwata and Ito 2011; Ishiwata et al. 2014, 2008; Kaeothip et al. 2013b). β-l-Arabinooligosaccharides were prepared according to previous reports (Fujita et al. 2014b). Other chemicals were purchased from Fujifilm Wako Pure Chemical Industries Ltd. (Osaka, Japan). Araf-β1,3-ArafGal2 was labeled with p-aminobenzoic ethyl ester (ABEE) as described by Wang (Wang et al. 1984). The ABEE derivative was purified on a Cosmosil PBr column with a linear gradient of CH3CN/water from 0:100 to 70:30 (v/v) for 30 min at a constant flow rate of 4.7 mL/min at 30 °C. Elution was monitored using a fluorescence detector (FP-2020, JASCO, Japan) with Ex/Em of 305/360 nm. Figure 1 depicts the chemical structures of the substrates used in this study.
Enzymes
β-l-Arabinofuranosidases Bll1HypBA1 (Fujita et al. 2014b), Bll2HypBA1, and Bll4HypBA1 (Ishiwata et al. 2023) enzymes were prepared according to previous reports. GH43_24 exo-β-1,3-galactanase Bl1,3Gal (Fujita et al. 2014a) and GH43_22 α-l-arabinofuranosidase BlArafA (Fujita et al. 2019a) were prepared as described previously. Endo-1,5-α-l-arabinanase (AnEARAB) and α-l-arabinofuranosidase (AnAFASE) from Aspergillus niger were purchased from Megazyme (Bray, Ireland). Table 1 shows the enzyme list used in this study.
Expression and purification of recombinant Bll3HypBA1
The genomic DNA of B. longum subsp. longum JCM 1217 was subjected to PCR amplification of the Bll3HypBA1 gene. The forward (5′-AGGAGATATACCATGGCAGAAGTGGACTCCAGCA-3′) and reverse (5′-TGCTCGAGTGCGGCCGCGTTCTGCATGCGCAC-3′) primers were designed using nucleotides 106–124 and 3655–3671 from the Bll3HypBA1 gene. The underlined texts indicate complementary nucleotides to the template. The PCR amplification of Bll3HypBA1-NΔ35CΔ761, which encodes amino acids (aa) 36–1223, was designed to eliminate the N-terminal signal peptide and C-terminal domains (Fig. 2A). The amplicon was cloned into the pET-23d vector (Novagen, Madison, WI, USA) using the In-Fusion HD Cloning Kit (Clontech Laboratories Inc., Palo Alto, CA, USA). The KOD-plus mutagenesis kit (Toyobo Co., Ltd., Osaka, Japan) and the primers listed in Table S1 were used to generate the Bll3HypBA1 deletion mutants. The primers NΔ reverse and NΔ379 forward were used to construct Bll3HypBA1-NΔ379CΔ761 (aa: 380–1223), whereas the primers CΔ forward and CΔ933 reverse were used to construct Bll3HypBA1-NΔ379CΔ933 (aa: 380–1051) (Fig. 2A). The constructed plasmids were sequenced and transformed into E. coli BL21 (λDE3) cells, which were then grown at 20 °C using the Overnight Express Autoinduction System (Novagen). The His-tagged proteins were extracted using BugBuster protein extraction reagent (Novagen), purified on a TALON metal affinity resin (Clontech Laboratories Inc.), and then desalted and concentrated using an ultrafiltration membrane (10-kDa cut-off; Millipore Co., Billerica, MA, USA). The sequence of the Bll3HypBA1-NΔ35CΔ761 gene was deposited in the DDBJ database under the accession number LC765464.
Sequence features of Bll3HypBA1 and GH127/146 β-l-arabinofuranosidases. A Domain structure of Bll3HypBA1. Domain structures were predicted using SignalP5.0 (https://services.healthtech.dtu.dk/services/SignalP-5.0/) and InterPro (https://www.ebi.ac.uk/interpro/) servers. The domains were indicated as follows: signal peptide (SP), laminin_G_3 (LamG), bacterial Ig-like domain (Ig), and transmembrane region (TM). Here, the lines indicate the expression regions of the Bll3HypBA1 recombinant proteins. B The phylogenetic tree of GH146 and GH127 β-l-arabinofuranosidase members in Bacteroides thetaiotaomicron VPI-5482, X. euvesicatoria, and B. longum subsp. longum JCM 1217. The phylogenetic tree was constructed with the MEGAX software. Enzymes characterized previously or in this study are enclosed in the box. Here, the asterisk (*) indicates enzymes for which crystallographic studies have been performed. Almost all enzymes were characterized as β-l-arabinofuranosidase, except for BT1003 (aceric acid hydrolase). GenBank accession numbers are shown alongside the characterized enzymatic names and/or protein locus tags as follows: Bll1HypBA1/BLLJ_0211 (BAJ65881), Bll2HypBA1/BLLJ_1826 (BAJ67491), Bll4HypBA1/BLLJ_0089 (BAJ65759), XeHypBA1/XCV2724 (CAJ24403), BT_0349 (AAO75456), BT_2911 (AAO78017), BT_0137 (AAO75244), BT3531 (AAO78637), BT2097 (AAO77204), BT1003 (AAO76110), BT3674 (AAO78779)
Thin-layer chromatography (TLC), high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD), and HPLC
For TLC, silica gel 60 aluminum plates (Merck KGaA, Darmstadt, Germany) were used with a 7:1:2 (v/v/v) of 1-propanol/EtOH/water solvent mixture and then visualized by spraying orcinol-sulfate on the plates (Holmes and O'Brien 1979). For HPAEC-PAD, oligosaccharides were analyzed using a CarboPac PA-1 column (Dionex Corp., Sunnyvale, CA, USA) as described previously (Sasaki et al. 2021). For HPLC, ABEE-labeled Araf-β1,3-ArafGal2 and reaction products were analyzed by a Cosmosil Sugar-D (Nacalai Tesque Inc.) as described previously (Ishiwata et al. 2022b).
pH and temperature dependence of Bll3HypBA1 activity
The hydrolytic activity of Bll3HypBA1-NΔ35CΔ761 was assayed using Araf-β1,3-ArafGal2-ABEE as the substrate and 50 mM sodium acetate (pH 3.5–6.0) and 50 mM sodium phosphate (pH 6.5–8.0) as buffers. The substrate (25 µM) was incubated with 0.05 µg/ml of Bll3HypBA1-NΔ35CΔ761 in 40 µL of each buffer (50 mM) at 40 °C for 20 min. The reaction was stopped by adding 10 µL of 5% trichloroacetic acid (TCA) and then analyzed via HPLC. Next, the effect of temperature on enzyme activity was examined using 50 mM sodium acetate buffer (pH 5.5) at 25–60 °C.
Substrate specificities of Bll3HypBA1 toward polysaccharides
The hydrolytic activity of Bll3HypBA1-NΔ35CΔ761 was analyzed using sugar beet arabinan, larch AG, and gum arabic AGP as polysaccharide substrates. These substrates (1.0%) were incubated with 0.1 µg/mL of Bll3HypBA1-NΔ35CΔ761, Bll1HypBA1, Bll2HypBA1, or Bll4HypBA1 in 40 µL of 50 mM sodium acetate buffer (pH 5.5) at 40 °C for 16 h. Following incubation, the released l-arabinose was analyzed using TLC, as described previously. For the comparison of the specific activity of Bll3HypBA1 with or without N-terminal LamG domain, the substrates (1.0%) were incubated with 0.1 µg/ml of Bll3HypBA1-NΔ35CΔ761 and Bll3HypBA1-NΔ379CΔ761 in 40 µL of 50 mM sodium acetate buffer (pH 5.5) at 40 °C for 2 h. The reaction was terminated by boiling, and the liberated l-arabinose was analyzed using HPAEC-PAD.
Combination reactions of Bll3HypBA1 and AGP degradative enzymes
Bll3HypBA1-NΔ35CΔ761 (0.1 µg/ml) reacted with Araf-β1,3-ArafGal2 and Araf-β1,3-Araf4 with or without BlArafA/AnAFASE at 40 °C for 16 h. The reaction mixtures were analyzed via HPAEC-PAD and MALDI-TOF MS. To determine the mode of action of rice AGP, the following reactions involving Bll3HypBA1-NΔ35CΔ761, BlArafA, and Bl1,3Gal were conducted: 1.0% rice AGP were incubated with enzymes (0.1 µg/mL each) in a 50 mM sodium phosphate buffer (pH 6.0) at 37 °C for 16 h. The reaction products were analyzed via HPAEC-PAD.
Substrate specificities of Bll3HypBA1 toward oligosaccharides
Next, the substrate specificities of Bll3HypBA1 were analyzed using synthetic and natural substrates. Since the region following residue 1052 in the NΔ379CΔ761 was not affected to structure of the catalytic region, Bll3HypBA1-NΔ379CΔ761 or CΔ933 were used for the analysis of substrate specificities. Synthetic Araf-β1,2-Araf-α-OMe, Araf-β1,3-Araf-α-OMe, and Araf-β1,5-Araf-α-OMe were incubated with 0.1 µg/ml of the Bll3HypBA1-NΔ379CΔ933 or Bll1HypBA1 in 40 µL of 50 mM sodium acetate buffer (pH 5.5) at 37 °C for 16 h. The reaction products were analyzed using TLC as previously described. To determine specific activities, natural oligosaccharides containing terminal Araf-β1,3 and Araf-β1,2 and synthetic Araf-β-pNP and Araf-β-OMe were used. Araf-β1,3-ArafGal2 (0.15 mM) and Araf-β1,3-Araf4 (0.15 mM) were incubated with 0.01 µg/mL of Bll3HypBA1-NΔ379CΔ761 in 40 µL of 50 mM sodium acetate buffer (pH 5.5) at 40 °C for 20 min. Araf-β-pNP (1.0 mM) was incubated with 0.1 µg/mL of Bll3HypBA1-NΔ379CΔ761 in 40 µL of 50 mM sodium acetate buffer (pH 5.5) at 40 °C for 2 h. A final concentration of 0.35 mM Araf-β1,2-Ara, 0.40 mM Araf-β-OMe, 0.11 mM Araf-β-Hyp (cis), 0.11 mM Araf-β-Hyp (trans), 0.13 mM Araf-β1,2-Araf-β-Hyp, and 1.0 mM Araf-β1,2-Araf-β1,2-Araf-β-Hyp were incubated with 12 µg/ml of the Bll3HypBA1-NΔ379CΔ761 in 40 µL of 50 mM sodium acetate buffer (pH 5.5) at 40 °C for 16 h. The reactions were terminated by adding 10 µL of 5% TCA and subsequently analyzed using HPAEC-PAD. One unit of enzyme activity was defined as the amount of enzyme required to produce 1 µmol l-arabinose per minute.
Transglycosylation activities
Transglycosylation reactions were performed using Arafβ1,3-AraGal2 as the donor and the 1-alkanols as acceptors. The substrate was incubated with 0.30 µg/mL of Bll3HypBA1-NΔ35CΔ761 in 40 µL of 50 mM sodium acetate buffer (pH 5.5) containing 5% methanol as the acceptor. After 1 h of incubation at 37 °C, the reaction was terminated by boiling. For ethanol and 1-propanol, the reactions were terminated after 3 h. TLC was used for confirmation of the reaction products as described above.
Protein purification for crystallization
For crystallization, selenomethionine (SeMet)-labeled and native proteins were expressed in E. coli BL21-CodonPlus (DE3)-RP-X and BL21-CodonPlus (DE3)-RIL (Agilent Technologies, Santa Clara, CA, USA), respectively. For the expression of the SeMet-labeled protein, the transformants were grown in Se-Met core medium supplemented with 10 g/L d-glucose, 250 mg/L MgSO4•7H2O, 4.2 mg/L FeSO4•7H2O, 10 ml/L Kao and Michayluk Vitamin Solution (Sigma-Aldrich), and 25 mg/L seleno-l-methionine. For the expression of the native protein, lysogeny broth was used. Transformants were grown at 37 °C for 2 h in a medium supplemented with 50 µg/mL chloramphenicol and 100 µg/mL ampicillin. Protein expression was induced by adding 0.1 mM isopropyl-β-d-thiogalactopyranoside and 0.5 mM ZnSO4 to the medium, and cells were further cultured at 18 °C for 24 h. Cells were harvested via centrifugation, suspended in 20 mM Tris–HCl (pH 7.5) and 300 mM NaCl (buffer A), and later disrupted via sonication; the supernatant was then purified via serial column chromatography. Ni-immobilized metal affinity chromatography was conducted using cOmplete™ His-Tag Purification Resin (Sigma-Aldrich) with wash and elution steps of 20 mM and 300 mM imidazole in buffer A, respectively. The buffer of the eluted protein sample was exchanged with 20 mM Tris–HCl (pH 7.5) (buffer B) using Vivaspin Turbo 15 (50-kDa cut-off; Sartorius Stedim Biotech, Göttingen, Germany). The protein sample was applied to a HiTrap Q HP column (5 mL) (Cytiva, Marlborough, MA, USA) equilibrated with buffer B and eluted with a linear gradient of 0 to 500 mM NaCl. Then, the sample was concentrated using Vivaspin Turbo 15 MWCO 50,000, and the solution was changed to 20 mM Tris–HCl (pH 7.5) and 150 mM NaCl (buffer C). Subsequently, gel filtration chromatography was conducted using a HiLoad 16/60 Superdex 200 pg column (Cytiva) equilibrated with buffer C at a flow rate of 1 mL/min. The purified protein was concentrated again using Vivaspin Turbo 15, and the solution was changed to buffer B. Protein concentrations were determined using a BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA) with bovine serum albumin as the standard.
Crystallography
The protein crystals were grown at 20 °C using the hanging drop vapor-diffusion method by combining the protein solution with an equal volume of a reservoir solution. SeMet-labeled Bll3HypBA1-NΔ379CΔ761 was crystallized using a 17 mg/mL protein solution and a reservoir solution containing 12% PEG 8000, 0.2 M MgCl2, and 0.1 M Tris–HCl (pH 8.5). The native Bll3HypBA1-NΔ379CΔ761 protein was crystallized using a protein solution (10 mg/mL) and a reservoir solution containing 15% PEG 8000, 0.2 M MgCl2, and 0.1 M Tris–HCl (pH 8.5). The crystals were cryoprotected in a reservoir solution supplemented with 25% glycerol. The native Bll3HypBA1-NΔ379CΔ933 protein was crystallized using a protein solution (14 mg/mL) and a reservoir solution containing 45% MPD, 0.2 M ammonium acetate, and 0.1 M HEPES–NaOH (pH 7.5). The crystal was soaked in a reservoir solution supplemented with 100 mM l-arabinose before cryocooling, but no electron density for l-arabinose was detected in the crystal structure. The crystals were flash-cooled by dipping into liquid nitrogen. X-ray diffraction data were collected at 100 K on beamlines at the Photon Factory of the High Energy Accelerator Research Organization (KEK, Tsukuba, Japan). Preliminary diffraction data were collected at SPring-8 (Hyogo, Japan). The datasets were processed using XDS (Kabsch 2010) and Aimless (Evans and Murshudov 2013). Phase determination and automated model building were performed for the data of SeMet-labeled crystal using the AutoSol pipeline of PHENIX (Adams et al. 2010). Manual model rebuilding and crystallographic refinement were performed using Coot (Emsley et al. 2010) and Refmac5 (Murshudov et al. 2011). Molecular graphic images were prepared using PyMOL (Schrödinger, LLC, New York, NY, USA).
Site-directed mutagenesis and activity assay
Site-directed mutants of Bll3HypBA1 were constructed using the PrimeSTAR mutagenesis basal kit (TaKaRa Bio, Ohtsu, Japan) with the primers shown in Table S1 and the pET23d-Bll3HypBA1-NΔ379CΔ933 plasmid as the template. These mutant enzymes were expressed and purified using the same procedures for Bll3HypBA1-NΔ379CΔ933. We also constructed a C805A mutant; however, it did not express in E. coli. The activity of the wild-type enzyme (NΔ379CΔ933) and mutants was assayed via TLC using Araf-β1,3-Araf-α-OMe as the substrate. The assay solution consisted of 12.5 mM Araf-β1,3-Araf-α-OMe, 0.0125 mg/mL enzyme, 10 mM dithiothreitol, and 20 mM sodium acetate buffer (pH 4.5). After incubation at 37 °C for 30 min, the reaction was stopped by heating the sample at 95 °C for 10 min. Then, 2 µL of the sample was spotted on a TLC plate (TLC Silica gel 60 F254, Merck) and developed using a 2:1:1 solvent mixture (v/v/v) of ethyl acetate/acetic acid/water. The separated sugars were visualized by spraying and heating with phosphomolybdic acid in ethanol on the plates. For mutants at Zn-coordinating and catalytic residues (E694Q, E694A, E723Q, E723A, C725S, C725A, C804S, C805S, and C805A) and E694Q/C804S double mutant, we determined specific activities via HPLC using Araf-β1,3-ArafGal2-ABEE as described above.
Results
Sequence features of Bll3HypBA1
Bll3HypBA1 (BLLJ_1848) contains a GH146 catalytic domain, a signal peptide, two laminin_G_3 (LamG) domains, which belong to the concanavalin A-like lectin/glucanase superfamily, three bacterial Ig-like domains, and a transmembrane domain (Fig. 2A). Furthermore, these features are shared with other cell surface anchoring glycosidases of B. longum subsp. longum JCM 1217, such as β-l-arabinobiosidase HypBA2 (BLLJ_0212) (Fujita et al. 2011), GH43 α-l-arabinofuranosidases HypAA (BLLJ_0213), BlArafA (BLLJ_1854) (Fujita et al. 2019a), BlArafB (BLLJ_1853), BlArafC (BLLJ_1852) (Komeno et al. 2022), BlArafD (BLLJ_1851) (Komeno et al. 2022), BlArafE (BLLJ_1850) (Sasaki et al. 2022), GH43_24 exo-β-1,3-galactanase Bl1,3Gal (BLLJ_1840) (Fujita et al. 2014a), and GH30_5 exo-β-1,6-galactobiohydrolase Bl1,6Gal (BLLJ_1841) (Fujita et al. 2019a). The aa sequence of the catalytic domain of Bll3HypBA1 exhibits 30% identity with the other GH146 paralog Bll4HypBA1, a β-l-arabinofuranosidase for Araf-β1-linked Hyp on the HRGP backbone (Ishiwata et al. 2023). In addition, Bll3HypBA1 exhibits 31% and 28% identities with the other characterized GH146 β-l-arabinofuranosidases BT0349 from Bacteroides thetaiotaomicron (Luis et al. 2018) and XeHypBA1 from X. euvesicatoria (Nakamura et al. 2018), respectively. Furthermore, the enzymes GH146 and GH127 are separated in the phylogenetic tree (Fig. 2B), and Bll3HypBA1 shows only 23% and 19% identities with the GH127 β-l-arabinofuranosidase paralogs Bll1HypBA1 and Bll2HypBA1 (BLLJ_1826), respectively. The alignment of the GH127 and GH146 catalytic domains revealed that the Zn-coordinating residues and catalytic residues are conserved in Bll3HypBA1 (Fig. S1).
Production of recombinant Bll3HypBA1 protein
The recombinant Bll3HypBA1-NΔ35CΔ761 protein without the N-terminal signal peptide and the C-terminal domains (Fig. 2A) was detected as a soluble protein only at a low expression level under induction condition at 20 °C. The purified recombinant Bll3HypBA1-NΔ35CΔ761 protein migrated as a single band on SDS-PAGE, which was in agreement with the theoretical molecular mass of 129,964 Da (Fig. S2). To obtain a clone suitable for X-ray crystallography and biochemical analysis, we constructed Bll3HypBA1-NΔ379CΔ761 and Bll3HypBA1-NΔ379CΔ933 without the LamG region. These clones were expressed as soluble proteins at 30 °C and purified as a single band in agreement with the theoretical molecular mass of 94,145 Da for Bll3HypBA1-NΔ379CΔ761 and 75,016 Da for Bll3HypBA1-NΔ379CΔ933 (Fig. S2). The estimated molecular masses by gel filtration chromatography were 91 kDa and 76 kDa for Bll3HypBA1-NΔ379CΔ761 and Bll3HypBA1-NΔ379CΔ933, respectively, suggesting that both protein constructs are monomeric in solution (data not shown).
Substrate specificity and general properties of the recombinant Bll3HypBA1 protein
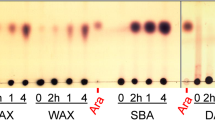
The substrate specificity of Bll3HypBA1 was compared with that of the other three paralogs of B. longum subsp. longum JCM 1217 using some natural polysaccharides as substrates. As shown in Fig. 3A, Bll3HypBA1-NΔ35CΔ761 released l-arabinose from sugar beet arabinan, larch AG, and gum arabic AGP, whereas other paralogs in B. longum subsp. longum JCM 1217 could not. This result suggests that Bll3HypBA1 has a specific ability to degrade the terminal β-l-Araf moieties of these polysaccharides. The l-arabinose content after the overnight reaction of Bll3HypBA1 was 0.32% for sugar beet arabinan, 0.39% for larch AG, and 0.20% for gum arabic AGP. This result indicates that a small amount of terminal β-l-Araf residues is present in AGPs and arabinans. Bll3HypBA1 also released l-arabinose from Araf-β1,3-ArafGal2 (Fig. 3B left). The mass of the degraded ArafGal2 was m/z 474.40 (calc. m/z 474.16), which was a reduction in the mass of l-arabinose (MW 132) from Araf-β1,3-ArafGal2 (m/z 606.48; calc. m/z 606.20). Subsequently, the released ArafGal2 was further hydrolyzed to β1,6-Gal2 by α-l-arabinofuranosidase BlArafA. Bll3HypBA1 also released l-arabinose from Araf-β1,3-Araf4 (m/z 678.77; calc. m/z 678.22), and the hydrolysate Araf4 was further degraded by A. niger α-l-arabinofuranosidase AnAFASE (Fig. 3B, right). The specific activity of Bll3HypBA1-NΔ379CΔ761 for Araf-β1,3-ArafGal2 was two times higher than that for Araf-β1,3-Araf4 (Table 2). In a comparison of the specific activities of larch AG, sugar beet arabinan, and gum arabic AGP, Bll3HypBA1-NΔ35CΔ761 with N-terminal LamG was 5.8-, 1.7-, and 1.1-fold higher than Bll3HypBA1-NΔ379CΔ761 without N-terminal LamG, respectively (Table S2). Alternatively, Bll3HypBA1-NΔ35CΔ761 was 1.6-fold lower than Bll3HypBA1-NΔ379CΔ761 for Araf-β1,3-ArafGal2.
Substrate specificities of Bll3HypBA1. A TLC analysis using sugar beet arabinan, larch AG, and gum arabic AGP as substrates. The substrates were incubated in the absence ( −) or presence ( +) of enzymes (Bll1HypBA1, Bll2HypBA1, Bll3HypBA1-NΔ35CΔ761, or Bll4HypBA1) at 37 °C for 16 h. B HPAEC-PAD analysis using Araf-β1,3-ArafGal2 and Araf-β1,3-Araf4 as substrates. The substrates were incubated with Bll3HypBA1-NΔ379CΔ761 at 40 °C for 16 h. The hydrolysate was further incubated with α-l-arabinofuranosidases BlArafA or AnAFASE. C TLC analysis using Araf-β1,2-Araf-α-OMe (β2), Araf-β1,3-Araf-α-OMe (β3), and Araf-β1,5-Araf-α-OMe (β5) in the absence ( −) or presence of Bll3HypBA1-NΔ379CΔ933 or Bll1HypBA1 at 37 °C for 16 h. aThe substrate was ( +) completely hydrolyzed, ( ±) partially hydrolyzed, and ( −) not hydrolyzed
Furthermore, synthetic arabinosyl disaccharides containing terminal β1,2-, β1,3-, and β1,5-Araf structures were used for a detailed evaluation of the linkage specificity of this enzyme (Ishiwata et al. 2022a). Bll3HypBA1 released l-arabinose from Araf-β1,3-Araf-α-OMe and slightly from Araf-β1,2-Araf-α-OMe but not from Araf-β1,5-Araf-α-OMe (Fig. 3C). Conversely, Bll1HypBA1 completely hydrolyzed these substrates. Bll3HypBA1 also exhibited < 1% degradative activity for Araf-β-pNP, Araf-β-OMe, Araf-β-Hyp glycosides, and β-AOSs containing Araf-β1,2-linkage compared with Araf-β1,3-ArafGal2 containing Araf-β1,3-linkage (Table 2). These results showed that Bll3HypBA1 is a β-l-arabinofuranosidase specific for Araf-β1,3-Araf structures.
The optimal temperature and pH for Araf-β1,3-ArafGal2-ABEE were 50 °C and 5.5, respectively (Fig. S3). Transglycosylation products were detected via TLC when 5% methanol, ethanol, and 1-propanol were used as acceptors (Fig. S4). These Rf values of TLC spots were similar to previous ones for Bll1HypBA1 (Fujita et al. 2014b). This result indicates that Bll3HypBA1 is the same anomer-retaining enzyme as other GH127/146 enzymes.
The rice anther AGP was predicted to contain Araf-β1,3-ArafGal2 side chains, as depicted in the schematic in Fig. 4’s right column. To clarify the importance of Bll3HypBA1 in the degradation of AGPs, a combination reaction with AGP-degrading enzymes was performed. The reaction of Bll3HypBA1 alone produced a small amount of l-arabinose, whereas the reaction of Bl1,3Gal and BlArafA without Bll3HypBA1 produced Araf-β1,3-ArafGal2 in addition to l-arabinose, galactose, and β1,6-Gal2 (Fig. 4). In contrast, Araf-β1,3-ArafGal2 was completely degraded to l-arabinose and β1,6-Gal2 in a combination reaction involving three enzymes. The releasing β1,6-Gal2 is probably degraded to galactose by intracellular GH42 β-galactosidase (BLLJ_0443) in B. longum subsp. longum JCM 1217 (Fujita et al. 2019b). These findings suggest that Bll3HypBA1 is an effective cell surface anchoring enzyme for the degradation of AGPs.
Stepwise hydrolysis of rice AGP by bifidobacterial AGP degradative enzymes. Rice AGP was incubated with a combination of Bl1,3Gal, BlArafA, and Bll3HypBA1-NΔ35CΔ761. A schematic drawing is shown in the right column. The cleavage sites of enzymes: Bl1,3Gal (black arrow), BlArafA (gray arrow), and Bll3HypBA1 (white arrow)
Crystal structure of Bll3HypBA1
The crystal structure of Bll3HypBA1-NΔ379CΔ761 (aa: 380–1223) was determined at a resolution of 1.75 Å by the single-wavelength anomalous dispersion using a SeMet-derivative (Table S3). The NΔ379CΔ761 structure contained one molecule per asymmetric unit, and residues M379–V1051 were modeled. In the active site of the NΔ379CΔ761 structure, a Zn atom coordinated by E723, C725, C804, and C805 was clearly observed, and a Tris molecule was bound to the l-arabinose-binding site (subsite − 1) (Fig. S5A and B). A crystallographic anomalous scattering analysis was performed to identify the metal atom (Table S3). A Bijvoet difference density map of the data collected at 1.280 Å wavelength revealed a prominent peak at the metal site, whereas a difference map of the data collected at 1.300 Å wavelength, which is above the absorption edge of Zn (1.2837 Å), showed a significant decrease in the anomalous scattering ability (Fig. S5C). This result confirmed that the metal ion bound at the active site is Zn2+.
Since the region following residue 1052 in the NΔ379CΔ761 structure was disordered, we also crystallized the Bll3HypBA1-NΔ379CΔ933 (aa: 380–1051) construct. The crystal structure of NΔ379CΔ933 was determined at a resolution of 1.70 Å (Table S3). The NΔ379CΔ933 structure contained two molecules per asymmetric unit, and residues from M379 to E1050 were modeled for both chains (A and B). The three molecules, including one in NΔ379CΔ761 and two in NΔ379CΔ933, had almost identical main chain structures, as the root mean square deviation (RMSD) values for the Cα atoms between all chain pairs were less than 0.24 Å. The Zn-coordination site structure of NΔ379CΔ933 was also almost identical to that of NΔ379CΔ761 (Fig. S5D), but no Tris molecule was identified in the active site. Therefore, we primarily focused on the chain A structure of NΔ379CΔ933.
The crystal structure of GH146 Bll3HypBA1 consists of three domains: catalytic (α/α)6 barrel (aa: 379–820), β-sandwich 1 (aa: 821–920), and β-sandwich 2 (aa: 921–1051) (Fig. 5A). The GH146 family member BT0349 has an α-helical linker and a jelly roll domain following the β-sandwich 2 domain (Fig. 5B). Although the C-terminal jelly roll domain of BT0349 is structurally similar to carbohydrate-binding module (CBM) family 35, the function of this domain remains unknown (Luis et al. 2018). While the GH146 enzymes (Bll3HypBA1 and BT0349) are monomeric in solution, GH127 Bll1HypBA1 forms a homodimer by interactions between the β-sandwich 2 domains (Fig. 5C). Extended structural elements in the β-sandwich 2 domain of the GH146 enzymes (a loop with two β-strands in Bll3HypBA1 and the jelly roll domain in BT0349) interact with the catalytic domain (Fig. 5A and B).
Overall and the catalytic domain structures of the GH146 and GH127 enzymes. A–C Domain structures of A GH146 Bll3HypBA1 and B GH146 BT0349 and dimer structure of C GH127 Bll1HypBA1. A ribbon presentation with a rainbow color (blue to red from the N- to C-terminus) is shown. Red circles indicate the Zn atoms at the active site (gray spheres). D–F Top views of the catalytic domain of D GH146 Bll3HypBA1, E GH146 BT0349, and F GH127 Bll1HypBA1. In E, an α-helix linker and the C-terminal jelly roll domain are shown in transparent color (orange to red)
Figure 5D, E, and F depict the top views of the catalytic domain of GH146 and GH127 enzymes. In Bll3HypBA1, loop 1 in β-sandwich 2 (aa: 947–969) partially covers the active site, whereas loop 2 (aa: 707–718) supports loop 1 (Fig. 5D). The C-terminal jelly roll domain of BT0349 shields its active site (Fig. 5E, transparent red). Loop 1 (aa: 566–590) is distant from the active site in BT0349, whereas loop 2 is absent. In GH127 Bll1HypBA1, the active site is covered by a long loop A (aa: 35–49) in the (α/α)6 barrel domain (Fig. 5F) (Ito et al. 2014).
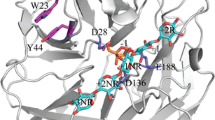
Figure 6 depicts the active site structures of the GH146 and GH127 enzymes. The Zn atom is coordinated by E723, C725, C804, and C805 in Bll3HypBA1 (Fig. 6A). The Zn-coordination structure of Bll3HypBA1 is identical to that of BT0349 and Bll1HypBA1 (Fig. 6B and C). R273, H194, H142, and Y145 form the substrate binding site of GH127 Bll1HypBA1 (Fig. 6C), whereas these residues are completely different in GH146 enzymes. F448 and I522 occupy the active site in Bll3HypBA1 (Fig. 6A), whereas W162 occupies this position in BT0349 (Fig. 6B). Therefore, these residues may determine the β-linkage specificity of Bll3HypBA1.
Mutational analysis
Next, using site-directed mutagenesis, we determined the significance of active site residues on enzyme activity (Fig. 7). Substitutions were made at the Zn-coordinating residues (E723, C725, C804, and C805), catalytic residues (E694 and C804), and residues near the catalytic center (W429, Y526, E578, H628, N630, T631, and Y771). The activity of the mutants was analyzed via TLC using Araf-β1,3-Araf-α-OMe as the substrate. The mutant enzymes at the Zn-coordinating and catalytic residues (E694Q, E694A, E723Q, E723A, C725S, C725A, C804S, C805S, and C805A) completely lost their activity (Fig. 7A). HPLC analysis also confirmed the importance of these residues (Table S4). For other active site residues, E578A also lost its activity, whereas W429A, Y526A, and N630A exhibited slight spots of l-arabinose release on TLC, indicating significantly diminished activity (Fig. 7A). H628A, T631A, and Y771A exhibited residual spots of the substrate Araf-β1,3-Araf-α-OMe but clear spots of l-arabinose after reaction at 37 °C for 30 min, indicating that these mutants retained their activity.
Mutational analysis of Bll3HypBA1. A TLC analysis of the wild-type Bll3HypBA1-NΔ379CΔ933 and its mutants using Araf-β1,3-Araf-α-OMe at 37 °C for 30 min. B Active site residues colored by the effects of mutation: magenta, no activity detected for mutants; green, weak activity detected by TLC; blue, activity detected; white, not examined
The result of the mutational analysis is shown in Fig. 7B by color codes. Residues without activity, residues with slight activity, and residues with significant activity in the mutant enzymes are depicted in magenta, green, and blue, respectively. The Zn-coordinating three Cys and one Glu residue and the acid/base catalyst residue were essential for activity. In the active site, E578 mutants lacked activity. W429 and E578 may participate in substrate recognition at subsite − 1 (Fig. 6A). N630 and Y526 play significant roles in the activity as well. H628, T631, and Y771 are involved in substrate recognition but are not required for activity. These nonessential residues may recognize the reducing end side of Araf-β1,3-Araf at subsite + 1.
Discussion
Previously, we characterized AGP-degrading enzymes in B. longum subsp. longum. The degradative enzymes are encoded by an AGP degradation gene cluster containing GH43_24 Bl1,3Gal (BLLJ_1840) and GH30_5 Bl1,6Gal (BLLJ_1841) and a GH43 gene cluster containing five GH43 α-l-arabinofuranosidases. GH43_22 BlArafA (BLLJ_1854) acts on the α1,3-linked Araf of AGP (Fujita et al. 2019a), GH43_22 BlArafB (BLLJ_1853) acts on the α1,5-linked Araf of the arabinan backbone, and GH43_27 BlArafC (BLLJ_1852) acts on the α1,2- and α1,3-linked Araf on the arabinan side chains (Komeno et al. 2019). Recently, BlArafD (BLLJ_1851) and BlArafE (BLLJ_1850) with tandem GH43 domains have been characterized. The GH43_UC (uncharacterized subfamily of GH43) domain of BlArafD acts on the α1,2-linked Araf of the α1,2- and α1,3-Araf doubly substituted arabinoxylan, whereas the GH43_26 BlArafD domain acts on the arabinan backbone (Komeno et al. 2022). In contrast, the GH43_22 BlArafE acts on the α1,3-linked Araf of the α1,3- and α1,4-Araf doubly substituted gum arabic AGP, whereas GH43_34 BlArafE acts on the remaining α1,4- linked Araf (Sasaki et al. 2022). A mixture of Bl1,3Gal, Bl1,6Gal, and BlArafA had a synergistic effect on the degradation of larch AG but only released 3.3% of the polysaccharide’s sugar component (Fujita et al. 2019a). A GH127 β-l-arabinofuranosidase, BT3674, in Bacteroides thetaiotaomicron showed a synergistic effect with exo-β-1,3-galactanases for the degradation of larch AG (Cartmell et al. 2018). Notably, Bll3HypBA1 (BLLJ_1848) is flanked by the AGP degradation gene cluster and GH43 gene cluster. In addition to Bll3HypBA1, all AGP degradation enzymes and GH43 α-l-arabinofuranosidases contain C-terminal membrane anchoring regions, indicating that these enzymes act synergistically to degrade AGPs and arabinans on the bifidobacterial cell surface. In contrast, the AGP degradation gene cluster is conserved in nearly all B. longum subsp. longum strains, whereas the Bll3HypBA1 ortholog (> 99% identity) was conserved in only 31 strains (5.2%) in 600 B. longum and B. longum subsp. longum strains in the NCBI genome database. In addition, the orthologous proteins (> 78% identity) are not conserved in other bifidobacterial species except for strains of B. aesculapii and B. primatium, which were isolated from the feces of common marmosets and cotton-top tamarins, respectively. These findings indicate that the Bll3HypBA1 ortholog is not a universal gene in bifidobacteria.
Bll3HypBA1 contains two independent LamG domain modules. Upon comparing the reactivities of Bll3HypBA1-NΔ35CΔ761 with N-terminal LamG and Bll3HypBA1-NΔ379CΔ761 without LamG, the former were more reactive for polysaccharides than the latter (Table S2). Notably, the N-terminal LamG domain was conserved in two adjacent AGP degradative enzymes (Bl1,3Gal and Bl1,6Gal) and four GH43 α-l-arabinofuranosidases (BlArafB-E) in B. longum subsp. longum JCM 1217. CBMs in bifidobacteria are predicted to serve as substrate and/or hydrolysate docking stations (van den Broek et al. 2008). The N-terminal LamG domain may aid in the degradation of AGPs and arabinans for bifidobacterial cell surface anchoring enzymes.
In the present study, the crystal structure of Bll3HypBA1 was determined as the second three-dimensional structure of the GH146 member following BT0349. BT0349 has a C-terminal jelly roll domain that covers the active site, and this enzyme does not appear to bind polymer substrates (Fig. 5B). BT0349 is present in the PUL of rhamnogalacturonan-I (RG-I), which comprises multiple GHs and is anticipated to act on oligosaccharides cleaved by other GHs. In contrast, Bll3HypBA1 can act independently on rice AGP (Fig. 4). This is consistent with the structural characteristics of Bll3HypBA1, which contains no large structural element covering the active site. The Zn atom of Bll3HypBA1 is coordinated by Cys3-Glu residues, and GH127 and GH146 enzymes share this structural characteristic (Cartmell et al. 2018; Ito et al. 2014; Luis et al. 2018; Maruyama et al. 2022; McGregor et al. 2021). In our previous studies, we elucidated the reaction mechanism of GH127 Bll1HypBA1 using synthetic inhibitors that were specifically designed for cysteine glycosidase (Ishiwata et al. 2022b; Maruyama et al. 2022). It has been demonstrated that the coordination of cysteine to Zn2+ is necessary for deglycosylation from the thioglycosyl intermediate, which is energetically unfavorable in the absence of metal coordination (McGregor et al. 2021). Consequently, this zinc coordination structure is shared by clan GH-P (GH127 and GH146) with cysteine as the catalytic nucleophilic residue. In addition, we performed a mutational analysis of the active site residues of Bll3HypBA1 and demonstrated the significance of these residues for the enzyme activity toward β1,3-linked Araf disaccharide. In addition to Zn-coordinating and catalytic residues, E578 was demonstrated to be essential for activity. Several other active site residues have been shown to contribute significantly to the enzyme activity, providing a structural basis for the strict recognition toward the Araf-β1,3-linkages.
Although it is known that gum arabic AGP from Acacia senegal is partially substituted by Gal-α1,3- or Arap-β1,3- at the Araf-α1,3-terminal sugar (Sasaki et al. 2021), the presence of Araf-β1,3 substitution has never been reported. Bifidobacterial GH39 3-O-α-d-galactosyl-α-l-arabinofuranosidase has been previously characterized for the release of Gal-α1,3-Araf disaccharide and 3-O-β-l-arabinopyranosyl-α-l-arabinofuranosidase for the release of Arap-β1,3-Araf disaccharide on AGP (Sasaki et al. 2021, 2023). The presence of β-Araf in gum arabic AGP and larch AG has been demonstrated, but the linkage positions remain unclear (Cartmell et al. 2018). In addition to quinoa arabinan (Wefers et al. 2014) and rice AGP (Kawaguchi et al. 1996), we found Araf-β1,3 substitution in larch AG, gum arabic AGP, and sugar beet arabinan. These results indicated that AGPs and arabinans may contain terminal Araf-β1,3-linkages. Since partially substituted sugars prevent enzymatic degradation of these polysaccharides by bacteria, the presence of Bll3HypBA1 and orthologous enzymes is advantageous for some Bifidobacterium and Bacteroides species with AGP-degrading ability. Jones et al. found GH39 α-l-(β-1,2)-arabinofuranobiosidase from rumen fungi; this enzyme releases Araf-β1,2-Araf disaccharide on sugar beet arabinan (Jones et al. 2017). To the best of our knowledge, Araf-β1,3-Araf disaccharide-releasing enzyme has not been discovered. Bll3HypBA1 is useful for glycan structure analysis, oligosaccharide preparation, and Araf-β1,3 content measurement due to its specificity for the Araf-β1,3-Araf structure.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. https://doi.org/10.1107/S0907444909052925
Cartmell A, Muñoz-Muñoz J, Briggs JA, Ndeh DA, Lowe EC, Baslé A, Terrapon N, Stott K, Heunis T, Gray J, Yu L, Dupree P, Fernandes PZ, Shah S, Williams SJ, Labourel A, Trost M, Henrissat B, Gilbert HJ (2018) A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation. Nat Microbiol 3:1314–1326. https://doi.org/10.1038/s41564-018-0258-8
Das D, Maiti S, Maiti TK, Islam SS (2013) A new arabinoxylan from green leaves of Litsea glutinosa (Lauraeae): structural and biological studies. Carbohydr Polym 92:1243–1248. https://doi.org/10.1016/j.carbpol.2012.10.052
Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. https://doi.org/10.1107/S0907444910007493
Evans PR, Murshudov GN (2013) How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69:1204–1214. https://doi.org/10.1107/S0907444913000061
Fujita K, Sakamoto S, Ono Y, Wakao M, Suda Y, Kitahara K, Suganuma T (2011) Molecular cloning and characterization of a β-l-arabinobiosidase in Bifidobacterium longum that belongs to a novel glycoside hydrolase family. J Biol Chem 286:5143–5150. https://doi.org/10.1074/jbc.M110.190512
Fujita K, Sakaguchi T, Sakamoto A, Shimokawa M, Kitahara K (2014a) Bifidobacterium longum subsp. longum exo-β-1,3-galactanase, an enzyme for the degradation of type II arabinogalactan. Appl Environ Microbiol 80:4577–4584. https://doi.org/10.1128/aem.00802-14
Fujita K, Takashi Y, Obuchi E, Kitahara K, Suganuma T (2014b) Characterization of a novel β-l-arabinofuranosidase in Bifidobacterium longum: functional elucidation of a DUF1680 protein family member. J Biol Chem 289:5240–5249. https://doi.org/10.1074/jbc.M111.248690
Fujita K, Sakamoto A, Kaneko S, Kotake T, Tsumuraya Y, Kitahara K (2019a) Degradative enzymes for type II arabinogalactan side chains in Bifidobacterium longum subsp. longum. Appl Microbiol Biotechnol 103:1299–1310. https://doi.org/10.1007/s00253-018-9566-4
Fujita K, Sasaki Y, Kitahara K (2019b) Degradation of plant arabinogalactan proteins by intestinal bacteria: characteristics and functions of the enzymes involved. Appl Microbiol Biotechnol 103:7451–7457. https://doi.org/10.1007/s00253-019-10049-0
Holmes EW, O’Brien JS (1979) Separation of glycoprotein-derived oligosaccharides by thin-layer chromatography. Anal Biochem 93:167–170. https://doi.org/10.1016/S0003-2697(79)80131-1
Ishiwata A, Ito Y (2011) Synthesis of docosasaccharide arabinan motif of mycobacterial cell wall. J Am Chem Soc 133:2275–2291. https://doi.org/10.1021/ja109932t
Ishiwata A, Munemura Y, Ito Y (2008) NAP ether mediated intramolecular aglycon delivery: a unified strategy for 1,2-cis-glycosylation. Eur J Org Chem 2008:4250–4263. https://doi.org/10.1002/ejoc.200800249
Ishiwata A, Kaeothip S, Takeda Y, Ito Y (2014) Synthesis of the highly glycosylated hydrophilic motif of extensins. Angew Chem Int Ed Engl 53:9812–9816. https://doi.org/10.1002/anie.201404904
Ishiwata A, Fujita K, Fushinobu S, Tanaka K, Ito Y (2022a) Synthesis of naturally occurring β-l-arabinofuranosyl-l-arabinofuranoside structures towards the substrate specificity evaluation of β-l-arabinofuranosidase. Bioorg Med Chem 68:116849. https://doi.org/10.1016/j.bmc.2022.116849
Ishiwata A, Narita S, Kimura K, Tanaka K, Fujita K, Fushinobu S, Ito Y (2022b) Mechanism-based inhibition of GH127/146 cysteine glycosidases by stereospecifically functionalized L-arabinofuranosides. Bioorg Med Chem 75:117054. https://doi.org/10.1016/j.bmc.2022.117054
Ishiwata A, Tsunomachi H, Kameyama K, Sophon K, Nakamura M, Kitahara K, Tanaka K, Ito Y, Fujita K (2023) Bifidobacterial GH146 β-L-arabinofuranosidase (Bll4HypBA1) as the last enzyme for the complete removal of oligoarabinofuranosides from hydroxyproline-rich glycoproteins. ChemBioChem 24:e202200637. https://doi.org/10.1002/cbic.202200637
Ito T, Saikawa K, Kim S, Fujita K, Ishiwata A, Kaeothip S, Arakawa T, Wakagi T, Beckham GT, Ito Y, Fushinobu S (2014) Crystal structure of glycoside hydrolase family 127 β-L-arabinofuranosidase from Bifidobacterium longum. Biochem Biophys Res Commun 447:32–37. https://doi.org/10.1016/j.bbrc.2014.03.096
Jones DR, Uddin MS, Gruninger RJ, Pham TTM, Thomas D, Boraston AB, Briggs J, Pluvinage B, McAllister TA, Forster RJ, Tsang A, Selinger LB, Abbott DW (2017) Discovery and characterization of family 39 glycoside hydrolases from rumen anaerobic fungi with polyspecific activity on rare arabinosyl substrates. J Biol Chem 292:12606–12620. https://doi.org/10.1074/jbc.M117.789008
Kabsch W (2010) Xds Acta Crystallogr D Biol Crystallogr 66:125–132. https://doi.org/10.1107/S0907444909047337
Kaeothip S, Ishiwata A, Ito T, Fushinobu S, Fujita K, Ito Y (2013a) Preparation of p-nitrophenyl β-l-arabinofuranoside as a substrate of β-l-arabinofuranosidase. Carbohydr Res 382:95–100. https://doi.org/10.1016/j.carres.2013.10.005
Kaeothip S, Ishiwata A, Ito Y (2013b) Stereoselective synthesis of Arabidopsis CLAVATA3 (CLV3) glycopeptide, unique protein post-translational modifications of secreted peptide hormone in plant. Org Biomol Chem 11:5892–5907. https://doi.org/10.1039/C3OB41212A
Kawaguchi K, Shibuya N, Ishii T (1996) A novel tetrasaccharide, with a structure similar to the terminal sequence of an arabinogalactan-protein, accumulates in rice anthers in a stage-specific manner. Plant J 9:777–785. https://doi.org/10.1046/j.1365-313X.1996.9060777.x
Komeno M, Hayamizu H, Fujita K, Ashida H (2019) Two novel α-l-arabinofuranosidases from Bifidobacterium longum subsp. longum belonging to glycoside hydrolase family 43 cooperatively degrade arabinan. Appl Environ Microbiol 85:e02582-e2618. https://doi.org/10.1128/AEM.02582-18
Komeno M, Yoshihara Y, Kawasaki J, Nabeshima W, Maeda K, Sasaki Y, Fujita K, Ashida H (2022) Two α-l-arabinofuranosidases from Bifidobacterium longum subsp. longum are involved in arabinoxylan utilization. Appl Microbiol Biotechnol 106:1957–1965. https://doi.org/10.1007/s00253-022-11845-x
Luis AS, Briggs J, Zhang X, Farnell B, Ndeh D, Labourel A, Baslé A, Cartmell A, Terrapon N, Stott K, Lowe EC, McLean R, Shearer K, Schückel J, Venditto I, Ralet M-C, Henrissat B, Martens EC, Mosimann SC, Abbott DW, Gilbert HJ (2018) Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol 3:210–219. https://doi.org/10.1038/s41564-017-0079-1
Maruyama S, Sawano K, Amaki S, Suzuki T, Narita S, Kimura K, Arakawa T, Yamada C, Ito Y, Dohmae N, Fujita K, Ishiwata A, Fushinobu S (2022) Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase. Glycobiology 32:171–180. https://doi.org/10.1093/glycob/cwab103
McGregor NGS, Coines J, Borlandelli V, Amaki S, Artola M, Nin-Hill A, Linzel D, Yamada C, Arakawa T, Ishiwata A, Ito Y, van der Marel GA, Codee JDC, Fushinobu S, Overkleeft HS, Rovira C, Davies GJ (2021) Cysteine nucleophiles in glycosidase catalysis: application of a covalent β-l-arabinofuranosidase inhibitor. Angew Chem Int Ed Engl 60:5754–5758. https://doi.org/10.1002/anie.202013920
Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. https://doi.org/10.1107/S0907444911001314
Nakamura M, Yasukawa Y, Furusawa A, Fuchiwaki T, Honda T, Okamura Y, Fujita K, Iwai H (2018) Functional characterization of unique enzymes in Xanthomonas euvesicatoria related to degradation of arabinofurano-oligosaccharides on hydroxyproline-rich glycoproteins. PLoS ONE 13:e0201982. https://doi.org/10.1371/journal.pone.0201982
Ndeh D, Rogowski A, Cartmell A, Luis AS, Basle A, Gray J, Venditto I, Briggs J, Zhang X, Labourel A, Terrapon N, Buffetto F, Nepogodiev S, Xiao Y, Field RA, Zhu Y, O’Neill MA, Urbanowicz BR, York WS, Davies GJ, Abbott DW, Ralet MC, Martens EC, Henrissat B, Gilbert HJ (2017) Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544:65–70. https://doi.org/10.1038/nature21725
Pellerin P, Doco T, Vidal S, Williams P, Brillouet JM, O’Neill MA (1996) Structural characterization of red wine rhamnogalacturonan II. Carbohydr Res 290:183–197
Ponder GR, Richards GN (1997) Arabinogalactan from Western larch, Part III: alkaline degradation revisited, with novel conclusions on molecular structure. Carbohydr Polym 34:251–261. https://doi.org/10.1016/S0144-8617(97)00099-4
Sasaki Y, Horigome A, Odamaki T, Xiao JZ, Ishiwata A, Ito Y, Kitahara K, Fujita K (2021) Novel 3-O-α-d-galactosyl-α-l-arabinofuranosidase for the assimilation of gum arabic arabinogalactan protein in Bifidobacterium longum subsp. longum. Appl Environ Microbiol 87:e02690-e2720. https://doi.org/10.1128/AEM.02690-20
Sasaki Y, Komeno M, Ishiwata A, Horigome A, Odamaki T, Xiao J-Z, Tanaka K, Ito Y, Kitahara K, Ashida H, Fujita K, Atomi H (2022) Mechanism of cooperative degradation of gum arabic arabinogalactan protein by Bifidobacterium longum surface enzymes. Appl Environ Microbiol 88:e02187-e2221. https://doi.org/10.1128/aem.02187-21
Sasaki Y, Yanagita M, Hashiguchi M, Horigome A, Xiao J-Z, Odamaki T, Kitahara K, Fujita K (2023) Assimilation of arabinogalactan side chains with novel 3-O-β-l-arabinopyranosyl-α-l-arabinofuranosidase in Bifidobacterium pseudocatenulatum. Microbiome Res Rep 2:12. https://doi.org/10.20517/mrr.2023.08
Tischer CA, Gorin PAJ, Iacomini M (2002) The free reducing oligosaccharides of gum arabic: aids for structural assignments in the polysaccharide. Carbohydr Polym 47:151–158. https://doi.org/10.1016/S0144-8617(01)00173-4
Tryfona T, Liang H-C, Kotake T, Kaneko S, Marsh J, Ichinose H, Lovegrove A, Tsumuraya Y, Shewry PR, Stephens E, Dupree P (2010) Carbohydrate structural analysis of wheat flour arabinogalactan protein. Carbohydr Res 345:2648–2656. https://doi.org/10.1016/j.carres.2010.09.018
van den Broek LA, Hinz SW, Beldman G, Vincken JP, Voragen AG (2008) Bifidobacterium carbohydrases-their role in breakdown and synthesis of (potential) prebiotics. Mol Nutr Food Res 52:146–163. https://doi.org/10.1002/mnfr.200700121
Wang WT, LeDonne NC Jr, Ackerman B, Sweeley CC (1984) Structural characterization of oligosaccharides by high-performance liquid chromatography, fast-atom bombardment-mass spectrometry, and exoglycosidase digestion. Anal Biochem 141:366–381. https://doi.org/10.1016/0003-2697(84)90057-5
Wefers D, Tyl CE, Bunzel M (2014) Novel arabinan and galactan oligosaccharides from dicotyledonous plants. Front Chem 2:1–9. https://doi.org/10.3389/fchem.2014.00100
Wefers D, Dong J, Abdel-Hamid AM, Paul HM, Pereira GV, Han Y, Dodd D, Baskaran R, Mayer B, Mackie RI, Cann I (2017) Enzymatic mechanism for arabinan degradation and transport in the thermophilic bacterium Caldanaerobius polysaccharolyticus. Appl Environ Microbiol 83:e00794-e817. https://doi.org/10.1128/AEM.00794-17
York WS, Kumar Kolli VS, Orlando R, Albersheim P, Darvill AG (1996) The structures of arabinoxyloglucans produced by solanaceous plants. Carbohydr Res 285:99–128. https://doi.org/10.1016/s0008-6215(96)90176-7
Acknowledgements
We thank Tomohide Shimotashiro (Kagoshima University, Japan) for the kind gifts of the young panicles of rice, Dr. Takatoshi Arakawa for helping experimental design, and the staff of Photon Factory and SPring-8 for the X-ray data collection.
Funding
Open access funding provided by Kagoshima University. This work was supported in part by JSPS KAKENHI Grant-in-Aid for Scientific Research (24580144 to K.F.; JP19H00929 to A.I.; JP18K05345 to A.I.; 15H02443 to S.F., K.F., and A.I.; 19H00929 to S.F., K.F., and A.I.; and JP 22H02196 to Y.I.) and Grant-in-Aid for Specially Promoted Research (JP16H06290 to Y.I. and A.I.).
Author information
Authors and Affiliations
Contributions
KF, AI, and SF conceived the project and designed the experiments. HT, PL, SM, MM, AD, KK, KT, YI, and AI conducted experiments. KF, AI, and SF drafted and edited the manuscript. All authors have reviewed the data and contributed to the completion of the manuscript.
Corresponding authors
Ethics declarations
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fujita, K., Tsunomachi, H., Lixia, P. et al. Bifidobacterial GH146 β-l-arabinofuranosidase for the removal of β1,3-l-arabinofuranosides on plant glycans. Appl Microbiol Biotechnol 108, 199 (2024). https://doi.org/10.1007/s00253-024-13014-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00253-024-13014-8