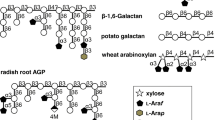
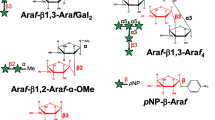
Abstract
Arabinoxylan (AX) and arabinoxylooligosaccharides (AXOs) are carbohydrate sources utilized by Bifidobacterium longum subsp. longum. However, their degradation pathways are poorly understood. In this study, we characterized two genes, BLLJ_1850 and BLLJ_1851, in the hemicellulose-degrading gene cluster (BLLJ_1836–BLLJ_1859) of B. longum subsp. longum JCM 1217. Both recombinant enzymes expressed in Escherichia coli exhibited exo-α-l-arabinofuranosidase activity toward p-nitrophenyl-α-l-arabinofuranoside. BlArafE (encoded by BLLJ_1850) contains the glycoside hydrolase family 43 (GH43), subfamily 22 (GH43_22), and GH43_34 domains. The BlArafE GH43_22 domain was demonstrated to release α1,3-linked Araf from AX, but the function of BlArafE GH43_34 could not be clearly identified in this study. BlArafD (encoded by BLLJ_1851) contains GH43 unclassified subfamily (GH43_UC) and GH43_26 domains. The BlArafD GH43_UC domain showed specificity for α1,2-linked Araf in α1,2- and α1,3-Araf double-substituted structures in AXOs, while BlArafD GH43_26 was shown to hydrolyze α1,5-linked Araf in the arabinan backbone. Co-incubation of BlArafD and BlArafE revealed that these two enzymes sequentially removed α1,2-Araf and α1,3-Araf from double-substituted AXOs in this order. B. longum strain lacking BLLJ_1850–BLLJ_1853 did not grow in the medium containing α1,2/3-Araf double-substituted AXOs, suggesting that BlArafE and BlArafD are important for the assimilation of AX.
Key points
• BlArafD GH43 unclassified subfamily domain is a novel α1,2- l -arabinofuranosidase.
• BlArafE GH43 subfamily 22 domain is an α1,3-l-arabinofuranosidase.
• BlArafD and BlArafE cooperatively degrade α1,2/3-Araf double-substituted arabinoxylan.
Graphical abstract







Similar content being viewed by others
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Amaretti A, Bernardi T, Leonardi A, Raimondi S, Zanoni S, Rossi M (2013) Fermentation of xylo-oligosaccharides by Bifidobacterium adolescentis DSMZ 18350: kinetics, metabolism, and β-xylosidase activities. Appl Microbiol Biotechnol 97:3109–3117. https://doi.org/10.1007/s00253-012-4509-y
Arboleya S, Bottacini F, O’Connell-Motherway M, Ryan CA, Ross RP, van Sinderen D, Stanton C (2018) Gene-trait matching across the Bifidobacterium longum pan-genome reveals considerable diversity in carbohydrate catabolism among human infant strains. BMC Genomics 19:33. https://doi.org/10.1186/s12864-017-4388-9
Ashida H, Maki R, Ozawa H, Tani Y, Kiyohara M, Fujita M, Imamura A, Ishida H, Kiso M, Yamamoto K (2008) Characterization of two different endo-α-N-acetylgalactosaminidases from probiotic and pathogenic enterobacteria, Bifidobacterium longum and Clostridium perfringens. Glycobiology 18:727–734. https://doi.org/10.1093/glycob/cwn053
Fujita K, Kitahara K, Suganuma T (2012) Functional analysis of degradative enzymes for hydroxyproline-linked β-l-arabinofuranosides in Bifidobacterium longum. Trends Glycosci Glycotechnol 24:215–224. https://doi.org/10.4052/tigg.24.215
Fujita K, Oura F, Nagamine N, Katayama T, Hiratake J, Sakata K, Kumagai H, Yamamoto K (2005) Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-α-N-acetylgalactosaminidase from Bifidobacterium longum. J Biol Chem 280:37415–37422. https://doi.org/10.1074/jbc.M506874200
Fujita K, Sakaguchi T, Sakamoto A, Shimokawa M, Kitahara K (2014) Bifidobacterium longum subsp. longum exo-β-1,3-galactanase, an enzyme for the degradation of type II arabinogalactan. Appl Environ Microbiol 80:4577–4584. https://doi.org/10.1128/AEM.00802-14
Fujita K, Sakamoto A, Kaneko S, Kotake T, Tsumuraya Y, Kitahara K (2019a) Degradative enzymes for type II arabinogalactan side chains in Bifidobacterium longum subsp. longum. Appl Microbiol Biotechnol 103:1299–1310. https://doi.org/10.1007/s00253-018-9566-4
Fujita K, Sasaki Y, Kitahara K (2019b) Degradation of plant arabinogalactan proteins by intestinal bacteria: characteristics and functions of the enzymes involved. Appl Microbiol Biotechnol 103:7451–7457. https://doi.org/10.1007/s00253-019-10049-0
Holck J, Lorentzen A, Vigsnæs LK, Licht TR, Mikkelsen JD, Meyer AS (2011) Feruloylated and nonferuloylated arabino-oligosaccharides from sugar beet pectin selectively stimulate the growth of Bifidobacterium spp. in human fecal in vitro fermentations. J Agric Food Chem 59:6511–6519. https://doi.org/10.1021/jf200996h
Ichinose H, Yoshida M, Fujimoto Z, Kaneko S (2008) Characterization of a modular enzyme of exo-1,5-α-l-arabinofuranosidase and arabinan binding module from Streptomyces avermitilis NBRC14893. Appl Microbiol Biotechnol 80:399–408. https://doi.org/10.1007/s00253-008-1551-x
Kitaoka M, Tian J, Nishimoto M (2005) Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl Environ Microbiol 71:3158–3162. https://doi.org/10.1128/AEM.71.6.3158-3162.2005
Komeno M, Hayamizu H, Fujita K, Ashida H (2019) Two novel α-l-arabinofuranosidases from Bifidobacterium longum subsp. longum belonging to glycoside hydrolase family 43 cooperatively degrade arabinan. Appl Environ Microbiol 85:e02582-e2618. https://doi.org/10.1128/AEM.02582-18
Mewis K, Lenfant N, Lombard V, Henrissat B (2016) Dividing the large glycoside hydrolase family 43 into subfamilies: a motivation for detailed enzyme characterization. Appl Environ Microbiol 82:1686–1692. https://doi.org/10.1128/AEM.03453-15
Michlmayr H, Hell J, Lorenz C, Böhmdorfer S, Rosenau T, Kneifel W (2013) Arabinoxylan oligosaccharide hydrolysis by family 43 and 51 glycosidases from Lactobacillus brevis DSM 20054. Appl Environ Microbiol 79:6747–6754. https://doi.org/10.1128/AEM.02130-13
Miyake M, Terada T, Shimokawa M, Sugimoto N, Arakawa T, Shimizu K, Igarashi K, Fujita K, Fushinobu S (2020) Structural analysis of β-l-arabinobiose-binding protein in the metabolic pathway of hydroxyproline-rich glycoproteins in Bifidobacterium longum. FEBS J 287:5114–5129. https://doi.org/10.1111/febs.15315
O’Callaghan A, van Sinderen D (2016) Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol 7:925. https://doi.org/10.3389/fmicb.2016.00925:925
Odamaki T, Bottacini F, Kato K, Mitsuyama E, Yoshida K, Horigome A, Xiao JZ, Van Sinderen D (2018) Genomic diversity and distribution of Bifidobacterium longum subsp. longum across the human lifespan. Sci Rep 8:85. https://doi.org/10.1038/s41598-017-18391-x
Rivière A, Moens F, Selak M, Maes D, Weckx S, De Vuyst L (2014) The ability of bifidobacteria to degrade arabinoxylan oligosaccharide constituents and derived oligosaccharides is strain dependent. Appl Environ Microbiol 80:204–217. https://doi.org/10.1128/AEM.02853-13
Saito K, Viborg AH, Sakamoto S, Arakawa T, Yamada C, Fujita K, Fushinobu S (2020a) Crystal structure of β-l-arabinobiosidase belonging to glycoside hydrolase family 121. PLoS ONE 15:e0231513. https://doi.org/10.1371/journal.pone.0231513
Saito Y, Shigehisa A, Watanabe Y, Tsukuda N, Moriyama-Ohara K, Hara T, Matsumoto S, Tsuji H, Matsuki T (2020b) Multiple transporters and glycoside hydrolases are involved in arabinoxylan-derived oligosaccharide utilization in Bifidobacterium pseudocatenulatum. Appl Environ Microbiol 86:e01782-e1820. https://doi.org/10.1128/AEM.01782-20
Sakka M, Yamada K, Kitamura T, Kunitake E, Kimura T, Sakka K (2019) The modular arabinanolytic enzyme Abf43A-Abf43B-Abf43C from Ruminiclostridium josui consists of three GH43 modules classified in different subfamilies. Enzyme Microb Technol 124:23–31. https://doi.org/10.1016/j.enzmictec.2019.01.011
Sasaki Y, Komeno M, Ishiwata A, Horigome A, Odamaki T, Xiao J-Z, Tanaka K, Ito Y, Kitahara K, Ashida H, Fujita K (2022) Mechanism of cooperative degradative mechanism of gum arabic arabinogalactan protein by Bifidobacterium longum surface enzymes. Appl Environ Microbiol 88:e02187-e2221. https://doi.org/10.1128/aem.02187-21
Song AX, Li LQ, Yin JY, Chiou JC, Wu JY (2020) Mechanistic insights into the structure-dependant and strain-specific utilization of wheat arabinoxylan by Bifidobacterium longum. Carbohydr Polym 249:116886. https://doi.org/10.1016/j.carbpol.2020.116886
Van Den Broek LAM, Lloyd RM, Beldman G, Verdoes JC, McCleary BV, Voragen AGJ (2005) Cloning and characterization of arabinoxylan arabinofuranohydrolase-D3 (AXHd3) from Bifidobacterium adolescentis DSM20083. Appl Microbiol Biotechnol 67:641–647. https://doi.org/10.1007/s00253-004-1850-9
Yamada C, Gotoh A, Sakanaka M, Hattie M, Stubbs KA, Katayama-Ikegami A, Hirose J, Kurihara S, Arakawa T, Kitaoka M, Okuda S, Katayama T, Fushinobu S (2017) Molecular insight into evolution of symbiosis between breast-fed infants and a member of the human gut microbiome Bifidobacterium longum. Cell Chem Biol 24:515-524.e5. https://doi.org/10.1016/j.chembiol.2017.03.012
Acknowledgements
We thank Prof. Takane Katayama (Kyoto University, Japan) for providing E. coli BL21(λDE3)ΔlacZ.
Funding
This work was supported by JSPS KAKENHI grant number 18K05494 (to HA).
Author information
Authors and Affiliations
Contributions
HA and KF conceived and designed the study. MK, YY, JK, WN, KM, and YS conducted the experiments. MK, YY, JK, KF, and HA analyzed the data. MK and HA wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Komeno, M., Yoshihara, Y., Kawasaki, J. et al. Two α-l-arabinofuranosidases from Bifidobacterium longum subsp. longum are involved in arabinoxylan utilization. Appl Microbiol Biotechnol 106, 1957–1965 (2022). https://doi.org/10.1007/s00253-022-11845-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-11845-x