Abstract
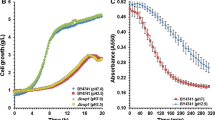
Kluyveromyces marxianus is a non-conventional yeast with outstanding physiological characteristics and a high potential for lignocellulosic ethanol production. However, achieving high ethanol productivity requires overcoming several biotechnological challenges due to the cellular inhibition caused by the inhibitors present in the medium. In this work, K. marxianus SLP1 was adapted to increase its tolerance to a mix of inhibitory compounds using the adaptive laboratory evolution strategy to study the adaptation and stress response mechanisms used by this non-Saccharomyces yeast. The fermentative and physiological parameters demonstrated that the adapted K. marxianus P8 had a better response against the synergistic effects of multiple inhibitors because it reduced the lag phase from 12 to 4 h, increasing the biomass by 40% and improving the volumetric ethanol productivity 16-fold than the parental K. marxianus SLP1. To reveal the effect of adaptation process in P8, transcriptome analysis was carried out; the result showed that the basal gene expression in P8 changed, suggesting the biological capability of K. marxianus to activate the adaptative prediction mechanism. Similarly, we carried out physiologic and transcriptome analyses to reveal the mechanisms involved in the stress response triggered by furfural, the most potent inhibitor in K. marxianus. Stress response studies demonstrated that P8 had a better physiologic response than SLP1, since key genes related to furfural transformation (ALD4 and ALD6) and stress response (STL1) were upregulated. Our study demonstrates the rapid adaptability of K. marxianus to stressful environments, making this yeast a promising candidate to produce lignocellulosic ethanol.
Key points
• K. marxianus was adapted to increase its tolerance to a mix of inhibitory compounds
• The basal gene expression of K. marxianus changed after the adaptation process
• Adapted K. marxianus showed a better physiological response to stress by inhibitors
• Transcriptome analyses revealed key genes involved in the stress response








Similar content being viewed by others
Data availability
The RNA-seq data and expression profiles were deposited in the Gene Expression Omnibus (GEO) repository belonging to NCBI under the bioproject PRJNA714250. The accession number of the K. marxianus SLP1 and P8 reads and their replicates is GSE176040. The expression profiles and metadata files are provided in the same bioproject.
Code availability
Not applicable.
References
Alcázar M, Kind T, Gschaedler A, Silveria M, Arrizon J, Fiehn O, Vallejo A, Higuera I, Lugo E (2017) Effect of steroidal saponins from agave on the polysaccharide cell wall composition of Saccharomyces cerevisiae and Kluyveromyces marxianus. LWT Food Sci Tech 77:430. https://doi.org/10.1016/j.lwt.2016.11.048
Allen SA, Clark W, Mccaffery JM, Cai Z, Lanctot A, Slininger PJ, Liu ZL, Gorsich SW (2010) Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels 3:1–10
Ayer A, Fellermeier S, Fife C, Li SS, Smits G, Meyer AJ, Dawes IW, Perrone GG (2012) A genome-wide screen in yeast identifies specific oxidative stress genes required for the maintenance of sub-cellular redox homeostasis. PLoS ONE 7(9):e44278. https://doi.org/10.1371/journal.pone.0044278
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477. https://doi.org/10.1089/cmb.2012.0021
Baptista M, Cunha JT, Domingues L (2021a) Establishment of Kluyveromyces marxianus as a microbial cell factory for lignocellulosic processes: production of high value furan derivatives. J Fungi 7(12):1047. https://doi.org/10.3390/jof7121047
Baptista SL, Costa CE, Cunha JT, Soares PO, Domingues L (2021b) Metabolic engineering of Saccharomyces cerevisiae for the production of top value chemicals from biorefinery carbohydrates. Biotechnol Adv 47:107697. https://doi.org/10.1016/j.biotechadv.2021.107697
Ben MZ, Khalil Y, Hersen P, Fabre E (2019) Hyperosmotic stress response memory is modulated by gene positioning in yeast. Cells 8:1–14. https://doi.org/10.3390/cells8060582
Bravim F, Mota MM, Fernandes AAR, Fernandes PMB (2016) High hydrostatic pressure leads to free radicals accumulation in yeast cells triggering oxidative stress. FEMS Yeast Res 16.https://doi.org/10.1093/femsyr/fow052
Celton M, Goelzer A, Camarasa C, Fromion V, Dequin S (2012) A constraint-based model analysis of the metabolic consequences of increased NADPH oxidation in Saccharomyces cerevisiae. Metab Eng 14:366–379. https://doi.org/10.1016/j.ymben.2012.03.008
Cunha JT, Romaní A, Costa CE, Sá-Correia I, Domingues L (2019) Molecular and physiological basis of Saccharomyces cerevisiae tolerance to adverse lignocellulose-based process conditions. Appl Microbiol Biotechnol 103:159–175. https://doi.org/10.1007/s00253-018-9478-3
Cunha JT, Soares PO, Baptista SL, Costa CE, Domingues L (2020) Engineered Saccharomyces cerevisiae for lignocellulosic valorization: a review and perspectives on bioethanol production. J Bioeng 11(1), 883–903. DOI: 10.1080/21655979.2020.1801178De Lomana ALG, Kaur A, Turkarslan S, Beer KD, Mast FD, Smith JJ, Aitchison JD, Baliga NS (2017) Adaptive prediction emerges over short evolutionary time scales. Genome Biol Evol 9:1616–1623. https://doi.org/10.1093/gbe/evx116
Dhar R, Sägesser R, Weikert C, Wagner A (2013) Yeast adapts to a changing stressful environment by evolving cross-protection and anticipatory gene regulation. Mol Biol Evol 30:573–588. https://doi.org/10.1093/molbev/mss253
Ding MZ, Wang X, Liu W, Cheng JS, Yang Y, Yuan YJ (2012) Proteomic research reveals the stress response and detoxification of yeast to combined inhibitors. PLoS ONE 7(8):e43474. https://doi.org/10.1371/journal.pone.0043474
Du C, Li Y, Xiang R, He Y, Sun H, Wang H, Yuan W (2022) Co-utilization of multiple lignocellulose-derived sugars and inhibitors by the robust mutant Kluyveromyces marxianus through adaptive laboratory evolution. Fuel 315:122816. https://doi.org/10.1016/j.fuel.2021.122816
Fabrizio P, Garvis S, Palladino F (2019) Histone methylation and memory of environmental stress. Cells 8:339. https://doi.org/10.3390/cells8040339
Flores-Cosío G, Arellano-Plaza M, Gschaedler A, Amaya-Delgado L (2018) Physiological response to furan derivatives stress by Kluyveromyces marxianus SLP1 in ethanol production. Rev Mex Ing Quim 17:189–202. https://doi.org/10.24275/uam/izt/dcbi/revmexingquim/2018v17n1/Flores
Flores-Cosío G, Herrera-López EJ, Arellano-Plaza M, Gschaedler-Mathis A, Sanchez A, Amaya-Delgado L (2019) Dielectric property measurements as a method to determine the physiological state of Kluyveromyces marxianus and Saccharomyces cerevisiae stressed with furan aldehydes. Appl Microbiol Biotechnol 103:9633–9642. https://doi.org/10.1007/s00253-019-10152-2
Freitas C, Neves E, Reis A, Passarinho PC, Lopes T (2013) Use of multi-parameter flow cytometry as tool to monitor the impact of formic acid on Saccharomyces carlsbergensis batch ethanol fermentations. Appl Biochem Biotechnol 169:2038–2048. https://doi.org/10.1007/s12010-012-0055-4
Fu X, Li P, Zhang L, Li S (2019) Understanding the stress responses of Kluyveromyces marxianus after an arrest during high-temperature ethanol fermentation based on integration of RNA-Seq and metabolite data. Appl Microbiol Biotechnol 103:2715–2729. https://doi.org/10.1007/s00253-019-09637-x
Gao J, Yuan W, Li Y, Bai F, Jiang Y (2017) Synergistic effect of thioredoxin and its reductase from Kluyveromyces marxianus on enhanced tolerance to multiple lignocellulose-derived inhibitors. Microb Cell Factories 16:1–13. https://doi.org/10.1186/s12934-017-0795-5
Gemayel R, Yang Y, Dzialo MC, Kominek J, Vowinckel J, Saels V, van Huffel L, van der Zande E, Ralser M, Steensels J, Voordeckers K, Verstrepen KJ (2017) Variable repeats in the eukaryotic polyubiquitin gene ubi4 modulate proteostasis and stress survival. Nat Commun. https://doi.org/10.1038/s41467-017-00533-4
Giannattasio S, Guaragnella N, Ždralevi M (2013) Molecular mechanisms of Saccharomyces cerevisiae stress adaptation and programmed cell death in response to acetic acid. Front Microbiol 4:1–7. https://doi.org/10.3389/fmicb.2013.00033
Gibney PA, Schieler A, Chen JC, Rabinowitz JD, Botstein D (2015) Characterizing the in vivo role of trehalose in Saccharomyces cerevisiae using the AGT1 transporter. Proc Natl Acad Sci USA 112:6122–6127. https://doi.org/10.1073/pnas.1506289112
Godinho CP, Prata CS, Pinto SN, Cardoso C, Narcisa MB (2018) Pdr18 is involved in yeast response to acetic acid stress counteracting the decrease of plasma membrane ergosterol content and order. Sci Rep 8:1–13. https://doi.org/10.1038/s41598-018-26128-7
Gómez-Márquez C, Sandoval-Nuñez D, Gschaedler A, Romero-Gutiérrez T, Amaya-Delgado L, Morales JA (2022) Diploid genome assembly of Kluyveromyces marxianus NRRL Y-50883 (SLP1). G3-Genes Genom Genet 12:1–6. https://doi.org/10.1093/g3journal/jkab347
Gu H, Zhang J, Bao J (2014) Inhibitor analysis and adaptive evolution of Saccharomyces cerevisiae for simultaneous saccharification and ethanol fermentation from industrial waste corncob residues. Bioresour Technol 157:6–13. https://doi.org/10.1016/j.biortech.2014.01.060
Hemansi, Himanshu, Patel AK, Saini JK, Singhania RR (2022) Development of multiple inhibitor tolerant yeast via adaptive laboratory evolution for sustainable bioethanol production. Bioresour Technol 344: Part B: 126247. https://doi.org/10.1016/j.biortech.2021.126247
Ikner A, Shiozaki K (2005) Yeast signaling pathways in the oxidative stress response. Mutation Research - Mutant Res-Fund Mol M 569:13–27. https://doi.org/10.1016/j.mrfmmm.2004.09.006
Jönsson LJ, Martín C (2016) Pretreatment of lignocellulose: formation of inhibitory by-products and strategies for minimizing their effects. Bioresour Technol 199:103–112. https://doi.org/10.1016/j.biortech.2015.10.009
Kahr H, Helmberger S, Jäger AG (2011) Yeast adaptation on the substrate straw. World renewable energy congress May 8–13, 2011: Linköping, Sweden, 492–499.
Kim D, Hahn JS (2013) Roles of the Yap1 transcription factor and antioxidants in Saccharomyces cerevisiae’s tolerance to furfural and 5-hydroxymethylfurfural, which function as thiol-reactive electrophiles generating oxidative stress. Appl Environ Microbiol 79:5069–5077. https://doi.org/10.1128/AEM.00643-13
Ko JK, Lee S (2018) Advances in cellulosic conversion to fuels: engineering yeasts for cellulosic bioethanol and biodiesel production. Curr Opin Biotechnol 50:72–80. https://doi.org/10.1016/j.copbio.2017.11.007
Kroon A, Rijken PJ, Smet CH (2013) Checks and balances in membrane phospholipid class and acyl chain homeostasis, the yeast perspective. Prog Lipid Res. 52:374–394. https://doi.org/10.1016/j.plipres.2013.04.006
Láinez M, Ruiz HA, Arellano-Plaza M, Martínez-Hernández S (2019) Bioethanol production from enzymatic hydrolysates of Agave salmiana leaves comparing S. cerevisiae and K. marxianus. Renew Energy 138:1127–1133. https://doi.org/10.1016/j.renene.2019.02
Lane M, Morrisey JP (2010) Kluyveromyces marxianus: a yeast emerging from its sister´s shadow. Fungal Biol Rev 24:17–26
Li X, Xiao ZT, Tan YL, Pei YP, Wang SJ, Feng XN, Wang QY, Zhang XL, Ye H, Li JH, Luo YD, Zou K, Shen XL (2022) Transcriptomic analysis reveals process of autolysis of Kluyveromyces marxianus in vacuum negative pressure and the higher temperature. Int Microbiol. https://doi.org/10.1007/s10123-022-00240-1
Li P, Fu X, Chen M, Zhang L, Li S (2019) Biotechnology for biofuels proteomic profiling and integrated analysis with transcriptomic data bring new insights in the stress responses of Kluyveromyces marxianus after an arrest during high temperature ethanol fermentation. Biotechnol Biofuels 1–13. https://doi.org/10.1186/s13068-019-1390-2
Liu ZL (2021) Reasons for 2-furaldehyde and 5-hydroxymethyl-2-furaldehyde resistance in Saccharomyces cerevisiae: current state of knowledge and perspectives for further improvements. Appl Microbiol Biotechnol 105:2991–3007. https://doi.org/10.1007/s00253-021-11256-4/Published
Lopes T, Baptista C, Reis A, Passarinho PC (2017) Using flow cytometry to evaluate the stress physiological response of the yeast Saccharomyces carlsbergensis ATCC 6269 to the presence of 5-hydroxymethylfurfural during ethanol fermentations. Appl Biochem Biotechnol 181:1096–1107. https://doi.org/10.1007/s12010-016-2271-9
López-Martínez G, Rodríguez B, Margalef-Català M, Cordero-Otero R (2012) The STF2p hydrophilin from Saccharomyces cerevisiae is required for dehydration stress tolerance. PLoS ONE 7(3):e33324. https://doi.org/10.1371/journal.pone.0033324
Martín C, Marcet M, Almazán O, Jönsson LJ (2007) Adaptation of a recombinant xylose-utilizing Saccharomyces cerevisiae strain to a sugarcane bagasse hydrolysate with high content of fermentation inhibitors. Bioresour Technol 98:1767–1773. https://doi.org/10.1016/j.biortech.2006.07.021
Ming M, Wang X, Lian L, Zhang H, Gao W, Zhu B, Lou D (2019) Metabolic responses of: Saccharomyces cerevisiae to ethanol stress using gas chromatography-mass spectrometry. Mol Omics 15:216–221. https://doi.org/10.1039/c9mo00055k
Mo W, Wang M, Zhan R, Yu Y, He Y, Lu H (2019) Kluyveromyces marxianus developing ethanol tolerance during adaptive evolution with significant improvements of multiple pathways. Biotechnol Biofuels 12:1–15. https://doi.org/10.1186/s13068-019-1393-z
Modig T, Lide NG, Taherzadeh MJ (2002) Inhibition effects of furfural on alcohol dehydrogenase, aldehyde dehydrogenase and pyruvate dehydrogenase. Biochem 363:769–776. https://doi.org/10.1042/bj3630769
Morano KA, Grant CM, Moye-Rowley WS (2012) The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190:1157–1195. https://doi.org/10.1534/genetics.111.128033
Narayanan V, Sànchez INV, van Niel EWJ, Gorwa-Grauslund MF (2016) Adaptation to low pH and lignocellulosic inhibitors resulting in ethanolic fermentation and growth of Saccharomyces cerevisiae. AMB Express 6:59. https://doi.org/10.1186/s13568-016-0234-8
Nitiyon S, Keo C, Murata M, Lertwattanasakul N, Limtong S (2016) Efficient conversion of xylose to ethanol by stress-tolerant Kluyveromyces marxianus. Springerplus 5:185. https://doi.org/10.1186/s40064-016-1881-6
Palmer JM, Stajich J (2020). Funannotate v1.8.1: Eukaryotic genome annotation (v1.8.1). Zenodo. https://doi.org/10.5281/zenodo.4054262
Patro R, Duggal G, Love M, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. https://doi.org/10.1038/nmeth.4197
Pattanakittivorakul S, Tsuzuno T, Kosaka T, Murata M, Kanesaki Y, Yoshikawa H, Limtong S, Yamada M (2022) Evolutionary adaptation by repetitive long-term cultivation with gradual increase in temperature for acquiring multi-stress tolerance and high ethanol productivity in Kluyveromyces marxianus DMKU 3–1042. Microorganisms 10(4):798. https://doi.org/10.3390/microorganisms10040798
Peng H, He L, Haritos VS (2019) Flow-cytometry-based physiological characterization and transcriptome analyses reveal a mechanism for reduced cell viability in yeast engineered for increased lipid content. Biotechnol Biofuels 12:98. https://doi.org/10.1186/s13068-019-1435-6
Pilap W, Thanonkeo S, Klanrit P, Thanonkeo P (2018) The potential of the newly isolated thermotolerant Kluyveromyces marxianus for high-temperature ethanol production using sweet sorghum juice. 3 Biotech 8:1–10. https://doi.org/10.1007/s13205-018-1161-y
Pinheiro T, Lip KYF, García-Ríos E, Querol A, Teixeira J, van Gulik W, Guillamón JM, Domingues L (2020) Differential proteomic analysis by SWATH-MS unravels the most dominant mechanisms underlying yeast adaptation to non-optimal temperatures under anaerobic conditions. Sci Rep 10:22329. https://doi.org/10.1038/s41598-020-77846-w
Prasad RK, Chatterjee S, Mazumder PB, Gupta SK, Sharma S, Vairale MG, Datta S, Dwivedi SK, Gupta DK (2019) Bioethanol production from waste lignocelluloses: a review on microbial degradation potential. Chemosphere 231:588–606. https://doi.org/10.1016/j.chemosphere.2019.05.142
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. https://doi.org/10.1093/bioinformatics/btp616
Saini JK, Agrawal R, Satlewal A, Saini R, Gupta R, Mathur A, Tuli D (2015) Second generation bioethanol production at high gravity of pilot-scale pretreated wheat straw employing newly isolated thermotolerant yeast Kluyveromyces marxianus DBTIOC-35. RSC Adv 5:37485–37494. https://doi.org/10.1039/c5ra05792b
Sandoval-Nuñez D, Arellano-Plaza M, Gschaedler A, Arrizon J, Amaya-Delgado L (2018) A comparative study of lignocellulosic ethanol productivities by Kluyveromyces marxianus and Saccharomyces cerevisiae. Clean Technol Environ Policy 20:1491–1499. https://doi.org/10.1007/s10098-017-1470-6
Sharma NK, Behera S, Arora R, Kumar S (2016) Enhancement in xylose utilization using Kluyveromyces marxianus NIRE-K1 through evolutionary adaptation approach. Bioprocess Biosyst Eng 39:835–843. https://doi.org/10.1007/s00449-016-1563-3
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. https://doi.org/10.1093/bioinformatics/btv351
Smith J, van Rensburg E, Görgens JF (2014) Simultaneously improving xylose fermentation and tolerance to lignocellulosic inhibitors through evolutionary engineering of recombinant Saccharomyces cerevisiae harbouring xylose isomerase. BMC Biotech 14:1–17. https://doi.org/10.1186/1472-6750-14-41
Tapia H, Young L, Fox D, Bertozzi CR, Koshland D (2015) Increasing intracellular trehalose is sufficient to confer desiccation tolerance to Saccharomyces cerevisiae. Proc Natl Acad Sci USA 112:6122–6127. https://doi.org/10.1073/pnas.1506415112
Thi T, Nguyen M, Kitajima S, Izawa S (2014) Importance of glucose-6-phosphate dehydrogenase (G6PDH) for vanillin tolerance in Saccharomyces cerevisiae. J Biosci Bioeng 118:263–269. https://doi.org/10.1016/j.jbiosc.2014.02.025
Tomás-Pejó E, Ballesteros M, Oliva JM, Olsson L (2010) Adaptation of the xylose fermenting yeast Saccharomyces cerevisiae F12 for improving ethanol production in different fed-batch SSF processes. J Ind Microbiol Biotechnol 37:1211–1220. https://doi.org/10.1007/s10295-010-0768-8
Ueno K, Matsumoto Y, Uno J, Sasamoto K, Sekimizu K, Kinjo Y, Chibana H (2011) Intestinal resident yeast Candida glabrata requires Cyb2p-mediated lactate assimilation to adapt in mouse intestine. PLoS ONE 6(9):e24759. https://doi.org/10.1371/journal.pone.0024759
van Dijk M, Erdei B, Galbe M, Nygård Y, Olsson L (2019) Strain-dependent variance in short-term adaptation effects of two xylose-fermenting strains of Saccharomyces cerevisiae. Bioresour Technol 292:121922. https://doi.org/10.1016/j.biortech.2019.121922
Wang D, Wu D, Yang X, Hong J (2018) Transcriptomic analysis of thermotolerant yeast Kluyveromyces marxianus in multiple inhibitors tolerance. RSC Adv 8:14177–14192. https://doi.org/10.1039/C8RA00335A
Wilson WA, Roach PJ, Montero M, Baroja-Fernández E, Muñoz FJ, Eydallin G, Viale AM, Pozueta-Romero J (2010) Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol Rev 34:952–985. https://doi.org/10.1111/j.1574-6976.2010.00220.x
Zhang B, Zhang J, Wang D, Han R, Ding R, Gao X, Sun L, Hong J (2016) Simultaneous fermentation of glucose and xylose at elevated temperatures co-produces ethanol and xylitol through overexpression of a xylose-specific transporter in engineered Kluyveromyces marxianus. Bioresour Technol 216:227–237. https://doi.org/10.1016/j.biortech.2016.05.068
Zhang B, Ren L, Wang H, Xu D, Zeng X, Li F (2020) Glycerol uptake and synthesis systems contribute to the osmotic tolerance of Kluyveromyces marxianus. Enzyme Microb Technol 140.https://doi.org/10.1016/j.enzmictec.2020.109641
Acknowledgements
The author deeply appreciates the CONACYT-SENER and COECyTJAL for financial support for this study. Sandoval-Nuñez Dania received a grant from CONACYT, Mexico.
Funding
The author sincerely appreciated the financial support from CONACYT-SENER (Fondo de Sustentabilidad Energética, projects 245750 and 250014). Sandoval Nuñez Dania and Gómez Márquez Carolina received a scholarship from CONACYT, Mexico (scholarship number 436599 and 738133, respectively).
Author information
Authors and Affiliations
Contributions
LAD and AGM project conceptualization. TRG, CGM, and DSN data curation and formal transcriptomic analysis. LAD funding acquisition and project administration. DSN conducted experiments. LAD, DSN, and MAP designed the methodology. TRG and CGM analyzed the data from bioinformatic analysis. LAD and AGM: supervision and validation. DSN wrote the original draft. DSN, TRG, CGM, and LAD wrote reviews and editing. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sandoval-Nuñez, D., Romero-Gutiérrez, T., Gómez-Márquez, C. et al. Physiological and transcriptome analyses of Kluyveromyces marxianus reveal adaptive traits in stress response. Appl Microbiol Biotechnol 107, 1421–1438 (2023). https://doi.org/10.1007/s00253-022-12354-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-12354-7