Abstract
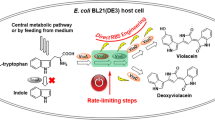
Violacein is a secondary metabolite mainly produced by Gram-negative bacteria that is formed from tryptophan by five enzymes encoded by a single operon. It is a broad-spectrum antibacterial pigment with various important biological activities such as anti-tumor, antiviral, and antioxidative effects. The newly discovered violacein operon vioABCDE was identified in the genome of the extremophile Janthinobacterium sp. B9-8. The key enzyme-encoding genes were cloned to construct the multigene coexpression plasmids pET-vioAB and pRSF-vioCDE. The violacein biosynthesis pathway was heterologously introduced into engineered Escherichia coli VioABCDE and VioABCDE-SD. The factors affecting violacein production, including temperature, pH, inoculum size, carbon and nitrogen source, precursor, and inducers were investigated. The violacein titer of VioABCDE-SD reached 107 mg/L in a two-stage fermentation process, representing a 454.4% increase over the original strain. The violacein operon from B9-8 provides a new microbial gene source for the analysis of the violacein synthesis mechanism, and the constructed engineering E. coli strains lay a foundation for the efficient and rapid synthesis of other natural products.
Key points
• The newly discovered violacein operon vioABCDE was identified in the genome of the extremophile Janthinobacterium sp. B9-8.
• The violacein synthesis pathway was reconstructed in E. coli using two compatible plasmids.
• A two-stage fermentation process was optimized for improved violacein accumulation.







Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information file).
References
Ahmad WA, Yusof NZ, Nordin N, Zakaria ZA, Rezali MF (2012) Production and characterization of violacein by locally isolated Chromobacterium violaceum grown in agricultural wastes. Appl Biochem Biotechnol 167(5):1220–1234
Ahmetagic A, Pemberton JM (2011) Antibiotic resistant mutants of Escherichia coli K12 show increases in heterologous gene expression. Plasmid 65(1):51–57
Alshatwi AA, Subash-Babu P, Antonisamy P (2016) Violacein induces apoptosis in human breast cancer cells through up regulation of BAX, p53 and down regulation of MDM2. Exp Toxicol Pathol 68(1):89–97
Boisbaudran L (1882) Matière colorante se formant dans la colle de farine. Comp Rend Acad Sci 94:562–563
Chen MH, Johns MR (1993) Effect of pH and nitrogen source on pigment production by Monascus purpureus. Appl Microbiol Biotechnol 40(1):132–138
Chen H, Bjerknes M, Kumar R, Jay E (1994) Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli m RNAs. Nucleic Acids Res 22(23):4953–4957
Choi SY, Kim S, Lyuck S, Kim SB, Mitchell RJ (2015) High-level production of violacein by the newly isolated Duganella violaceinigra str. NI28 and its impact on Staphylococcus aureus. Sci Rep 5(1):1–12
Choi KR, Cho JS, Cho IJ, Park D, Lee SY (2018) Markerless gene knockout and integration to express heterologous biosynthetic gene clusters in Pseudomonas putida. Metab Eng 47:463–474
Chu X, Liu J, Gu W, Tian L, Tang S, Zhang Z, Jiang L, Xu X (2021) Study of the properties of carotenoids and key carotenoid biosynthesis genes from Deinococcus xibeiensis R13. Biotechnol Appl Biochem. https://doi.org/10.1002/bab.2217
de Sousa Leal AM, de Queiroz JDF, de Medeiros SRB, de Souza Lima TK, Agnez-Lima LF (2015) Violacein induces cell death by triggering mitochondrial membrane hyperpolarization in vitro. BMC Microbiol 15(1):1–8
de Vasconcelos ATR, De Almeida DF, Hungria M, Guimaraes CT, Antônio RV, Almeida FC, De Almeida LG, De Almeida R, Alves-Gomes JA, Andrade EM (2003) The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc Natl Acad Sci U S A:11660–11665
DeMoss RD (1967) Violacein. Antibiotics 2:77–81
Durán N, Menck CF (2001) Chromobacterium violaceum: a review of pharmacological and industiral perspectives. Crit Rev Microbiol 27(3):201–222
Durán N, Justo GZ, Durán M, Brocchi M, Cordi L, Tasic L, Castro GR, Nakazato G (2016) Advances in Chromobacterium violaceum and properties of violacein-its main secondary metabolite: a review. Biotechnol Adv 34(5):1030–1045
Fang MY, Zhang C, Yang S, Cui JY, Jiang PX, Lou K, Wachi M, Xing XH (2015) High crude violacein production from glucose by Escherichia coli engineered with interactive control of tryptophan pathway and violacein biosynthetic pathway. Microb Cell Fact 14(1):1–13
Friedrich I, Hollensteiner J, Schneider D, Poehlein A, Hertel R, Daniel R (2020) First complete genome sequences of Janthinobacterium lividum EIF1 and EIF2 and their comparative genome analysis. Genome Biol Evol 12(10):1782–1788
Füller JJ, Röpke R, Krausze J, Rennhack KE, Daniel NP, Blankenfeldt W, Schulz S, Jahn D, Moser J (2016) Biosynthesis of violacein, structure and function of l-tryptophan oxidase VioA from Chromobacterium violaceum. J Biol Chem 291(38):20068–20084
Gahlout M, Chauhan PB, Prajapati H, Tandel N, Rana S, Solanki D, Patel N (2021) Characterization, application and statistical optimization approach for enhanced production of pyocyanin pigment by Pseudomonas aeruginosa DN9. Syst Microbiol and Biomanuf 1:459–470
Gwon DA, Seok JY, Jung GY, Lee JW (2021) Biosensor-assisted adaptive laboratory evolution for violacein production. Int J Mol Sci 22(12):6594
Hakobyan L, Gabrielyan L, Trchounian A (2012) Yeast extract as an effective nitrogen source stimulating cell growth and enhancing hydrogen photoproduction by Rhodobacter sphaeroides strains from mineral springs. Int J Hydrogen Energ 37(8):6519–6526
Hannig G, Makrides SC (1998) Strategies for optimizing heterologous protein expression in Escherichia coli. Trends Biotechnol 16(2):54–60
He L, Xiu Y, Jones JA, Baidoo EE, Keasling JD, Tang YJ, Koffas MA (2017) Deciphering flux adjustments of engineered E. coli cells during fermentation with changing growth conditions. Metab Eng 39:247–256
Helmetag V, Samel SA, Thomas MG, Marahiel MA, Essen LO (2009) Structural basis for the erythro-stereospecificity of the l-arginine oxygenase VioC in viomycin biosynthesis. FEBS J 276(13):3669–3682
Hirano S, Asamizu S, Onaka H, Shiro Y, Nagano S (2008) Crystal structure of VioE, a key player in the construction of the molecular skeleton of violacein. J Biol Chem 283(10):6459–6466
Immanuel SRC, Banerjee D, Rajankar MP, Raghunathan A (2018) Integrated constraints based analysis of an engineered violacein pathway in Escherichia coli. BioSyst 171:10–19
Jiang PX, Wang HS, Zhang C, Lou K, Xing XH (2010) Reconstruction of the violacein biosynthetic pathway from Duganella sp. B2 in different heterologous hosts. Appl Microbiol Biotechnol 86(4):1077–1088
Jin W, Xu X, Jiang L, Zhang Z, Li S, Huang H (2015) Putative carotenoid genes expressed under the regulation of Shine-Dalgarno regions in Escherichia coli for efficient lycopene production. Biotechnol Lett 37(11):2303–2310
Jones JA, Vernacchio VR, Lachance DM, Lebovich M, Fu L, Shirke AN, Schultz VL, Cress B, Linhardt RJ, Koffas MA (2015) ePathOptimize: a combinatorial approach for transcriptional balancing of metabolic pathways. Sci Rep 5(1):1–10
Kitcha S, Cheirsilp B (2013) Enhancing lipid production from crude glycerol by newly isolated oleaginous yeasts: strain selection, process optimization, and fed-batch strategy. Bioenerg Res 6(1):300–310
Kumar R, Acharya V, Singh D, Kumar S (2018) Strategies for high-altitude adaptation revealed from high-quality draft genome of non-violacein producing Janthinobacterium lividum ERGS5: 01. Stand Genomic Sci 13(1):1–13
Lamendella R, Jude BA (2018) Draft genome sequences of violacein-producing Duganella sp. isolates from a waterway in Eastern Pennsylvania. Microbiol Resour Ann 7(12):e01196-18
Lee ME, Aswani A, Han AS, Tomlin CJ, Dueber JE (2013) Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res 41(22):10668–10678
Li G-W, Oh E, Weissman JS (2012) The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484(7395):538–541
Lou J, Gu H, Wang H, An Q, Xu J (2016) Complete genome sequence of Massilia sp. WG5, an efficient phenanthrene-degrading bacterium from soil. J Biotechnol 218:49–50
Nappi J, Soldi E, Egan S (2019) Diversity and distribution of bacteria producing known secondary metabolites. Microb Ecol 78(4):885–894
Nguyen PH, Wu YY, Guo S, Murray RM (2015) Design space exploration of the violacein pathway in Escherichia coli based cell-free system (TX-TL). bioRxiv 027656:1–10. https://doi.org/10.1101/027656
Pantanella F, Berlutti F, Passariello C, Sarli S, Morea C, Schippa S (2007) Violacein and biofilm production in Janthinobacterium lividum. J Appl Microbiol 102(4):992–999
Park H, Park S, Yang Y-H, Choi K-Y (2021) Microbial synthesis of violacein pigment and its potential applications. Crit Rev Biotechnol 1–23
Pauer H, Hardoim CCP, Teixeira FL, Miranda KR, Barbirato DdS, Carvalho DPd, Antunes LCM, Leitão ÁAdC, Lobo LA, Domingues RMCP (2018) Impact of violacein from Chromobacterium violaceum on the mammalian gut microbiome. PLoS One 13(9):e0203748
Paulsen SS, Strube ML, Bech PK, Gram L, Sonnenschein EC (2019) Marine chitinolytic Pseudoalteromonas represents an untapped reservoir of bioactive potential. mSystems 4(4):e00060-19
Pemberton JM, Vincent KM, Penfold RJ (1991) Cloning and heterologous expression of the violacein biosynthesis gene cluster from Chromobacterium violaceum. Curr Microbiol 22(6):355–358
Ran T, Gao M, Wei Q, He J, Tang L, Wang W, Xu D (2015) Expression, crystallization and preliminary crystallographic data analysis of VioD, a hydroxylase in the violacein-biosynthesis pathway. Acta Crystallogr F 71(2):149–152
Raths R, Peta V, Bücking H (2019) Draft genome sequence of Duganella sp. strain DN04, isolated from cultivated soil. Microbiol Resour Ann 8(32):e00848-19
Rodrigues AL, Trachtmann N, Becker J, Lohanatha AF, Blotenberg J, Bolten CJ, Korneli C, de Souza Lima AO, Porto LM, Sprenger GA (2013) Systems metabolic engineering of Escherichia coli for production of the antitumor drugs violacein and deoxyviolacein. Metab Eng 20:29–41
Sarwar N, Sarwar S, Ejaz S, Al-Adeeb A, Al-Ansi W, Li Y, Bai Z (2021) Metabolic engineering of microorganisms to increase production of violacein. Int J Environ 6(1):295–306. https://doi.org/10.22161/ijeab.61.37
Segall-Shapiro TH, Sontag ED, Voigt CA (2018) Engineered promoters enable constant gene expression at any copy number in bacteria. Nat Biotechnol 36(4):352–358
Soby SD, Gadagkar SR, Contreras C, Caruso FL (2013) Chromobacterium vaccinii sp. nov., isolated from native and cultivated cranberry (Vaccinium macrocarpon Ait.) bogs and irrigation ponds. Int J Syst Evol Microbiol 63(5):1840–1846
Studier FW (2005) Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif 41(1):207–234
Sun H, Zhao D, Xiong B, Zhang C, Bi C (2016) Engineering Corynebacterium glutamicum for violacein hyper production. Microb Cell Fact 15(1):1–9
Thøgersen MS, Delpin MW, Melchiorsen J, Kilstrup M, Månsson M, Bunk B, Sproeer C, Overmann J, Nielsen KF, Gram L (2016) Production of the bioactive compounds violacein and indolmycin is conditional in a maeA mutant of Pseudoalteromonas luteoviolacea S4054 lacking the malic enzyme. Front Microbiol 7:1461
Tong Y, Zhou J, Zhang L, Xu P (2021) A golden-gate based cloning toolkit to build violacein pathway libraries in Yarrowia lipolytica. ACS Synth Biol 10(1):115–124
Wang H, Zhang X, Wang S, Zhao B, Lou K, Xing XH (2018) Massilia violaceinigra sp. nov., a novel purple-pigmented bacterium isolated from glacier permafrost. Int J Syst Evol Microbiol 68(7):2271–2278
Wong L, Holdridge B, Engel J, Xu P (2019) Genetic tools for streamlined and accelerated pathway engineering in Yarrowia lipolytica. Methods Mol Biol 1927:155–177
Xu X, Jin W, Jiang L, Xu Q, Li S, Zhang Z, Huang H (2016) A high-throughput screening method for identifying lycopene-overproducing E. coli strain based on an antioxidant capacity assay. Biochem Eng J 112:277–284
Xu J, Xu X, Xu Q, Zhang Z, Jiang L, Huang H (2018) Efficient production of lycopene by engineered E. coli strains harboring different types of plasmids. Bioproc Biosyst Eng 41(4):489–499
Xu X, Tian L, Zhang S, Jiang L, Zhang Z, Huang H (2019) Complete genome sequence of Janthinobacterium sp. B9–8, a violacein-producing bacterium isolated from low-temperature sewage. Microb Pathog 128:178–183. https://doi.org/10.1016/j.micpath.2019.01.003
Yang C, Jiang P, Xiao S, Zhang C, Lou K, Xing XH (2011) Fed-batch fermentation of recombinant Citrobacter freundii with expression of a violacein-synthesizing gene cluster for efficient violacein production from glycerol. Biochem Eng J 57:55–62
Zhang X, Enomoto K (2011) Characterization of a gene cluster and its putative promoter region for violacein biosynthesis in Pseudoalteromonas sp. 520P1. Appl Microbiol Biotechnol 90(6):1963–1971
Zhang Y, Chen H, Zhang Y, Yin H, Zhou C, Wang Y (2021) Direct RBS engineering of the biosynthetic gene cluster for efficient productivity of violaceins in E. coli. Microb Cell Fact 20(1):38. https://doi.org/10.1186/s12934-021-01518-1
Zhou Y, Fang MY, Li G, Zhang C, Xing XH (2018) Enhanced production of crude violacein from glucose in Escherichia coli by overexpression of rate-limiting key enzyme (s) involved in violacein biosynthesis. Appl Biochem Biotechnol 186(4):909–916
Funding
This work was supported by the National Natural Science Foundation of China (21978136, 32060004), the National Natural Science Foundation of China Youth Fund (31922070), and the Natural Science Foundation of Jiangsu Province (BK20211268, BK20180038).
Author information
Authors and Affiliations
Contributions
X. X. and L. J. designed the experiments. X. X., X. C., B. D., and C. H. performed the research. C. X. and Z. Z. collected and analyzed the data. Z. Z. and L. J. supervised the research. X. X. and X. C. wrote the paper. C. X., Z. Z., and L. J. polished the paper. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, X., Chu, X., Du, B. et al. Functional characterization of a novel violacein biosynthesis operon from Janthinobacterium sp. B9-8. Appl Microbiol Biotechnol 106, 2903–2916 (2022). https://doi.org/10.1007/s00253-022-11929-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-11929-8