Abstract
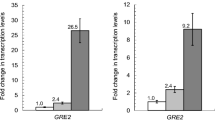
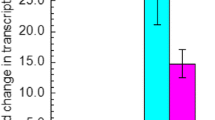
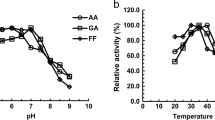
Aldehydes generated as by-products during the pretreatment of lignocellulose are the key inhibitors to Saccharomyces cerevisiae, which is considered as the most promising microorganism for industrial production of biofuel, xylitol as well as other special chemicals from lignocellulose. S. cerevisiae has the inherent ability to in situ detoxify aldehydes to corresponding alcohols by multiple aldehyde reductases. Herein, we report that an uncharacterized open reading frame YKL071W from S. cerevisiae encodes a novel “classical” short-chain dehydrogenase/reductase (SDR) protein with NADH-dependent enzymatic activities for reduction of furfural (FF), glycolaldehyde (GA), formaldehyde (FA), and benzaldehyde (BZA). This enzyme showed much better specific activities for reduction of GA and FF than FA and BZA, and displayed much higher Km and Kcat/Km but lower Vmax and Kcat for reduction of GA than FF. For this enzyme, the optimum pH was 5.5 and 6.0 for reduction of GA and FF, and the optimum temperature was 30 °C for reduction of GA and FF. Both pH and temperature affected stability of this enzyme in a similar trend for reduction of GA and FF. Cu2+, Zn2+, Ni2+, and Fe3+ had severe inhibition effects on enzyme activities of Ykl071wp for reduction of GA and FF. Transcription of YKL071W in S. cerevisiae was significantly upregulated under GA and FF stress conditions, and its transcription is most probably regulated by transcription factor genes of YAP1, CAD1, PDR3, and STB5. This research provides guidelines to identify more uncharacterized genes with reductase activities for detoxification of aldehydes derived from lignocellulose in S. cerevisiae.





Similar content being viewed by others
References
Agbor VB, Cicek N, Sparling R, Berlin A, Levin DB (2011) Biomass pretreatment: fundamentals toward application. Biotechnol Adv 29:675–685. https://doi.org/10.1016/j.biotechadv.2011.05.005
Allen SA, Clark W, McCaffery JM, Cai Z, Lanctot A, Slininger PJ, Liu ZL, Gorsich SW (2010) Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels 3:2. https://doi.org/10.1186/1754-6834-3-2
Bilal M, Asgher M, Iqbal HM, Hu H, Zhang X (2017) Biotransformation of lignocellulosic materials into value-added products—a review. Int J Biol Macromol 98:447–458. https://doi.org/10.1016/j.ijbiomac.2017.01.133
Cavka A, Stagge S, Jönsson LJ (2015) Identification of small aliphatic aldehydes in pretreated lignocellulosic feedstocks and evaluation of their inhibitory effects on yeast. J Agric Food Chem 63(44):9747–9754. https://doi.org/10.1021/acs.jafc.5b04803
Delgenes JP, Moletta R, Navarro JM (1996) Effects of lignocellulose degradation products on ethanol fermentations of glucose and xylose by Saccharomyces cerevisiae, Zymomonas mobilis, Pichia stipitis, and Candida shehatae. Enzym Microb Technol 19:220–225. https://doi.org/10.1016/0141-0229(95)00237-5
Engel SR, Dietrich FS, Fisk DG, Binkley G, Balakrishnan R, Costanzo MC, Dwight SS, Hitz BC, Karra K, Nash RS, Weng S, Wong ED, Lloyd P, Skrzypek MS, Miyasato SR, Simison M, Cherry JM (2014) The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 (Bethesda) 4(3):389–398. https://doi.org/10.1534/g3.113.008995
Gietz RD, Schiestl RH, Willems AR, Woods RA (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11(4):355–360. https://doi.org/10.1002/yea.320110408
Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG (1996) Life with 6000 genes. Science 274(5287):546, 563–546, 567. https://doi.org/10.1126/science.274.5287.546
Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24(13):2519–2524. https://doi.org/10.1093/nar/24.13.2519
Gulshan K, Rovinsky SA, Coleman ST, Moye-Rowley WS (2005) Oxidant-specific folding of Yap1p regulates both transcriptional activation and nuclear localization. J Biol Chem 280(49):40524–40533. https://doi.org/10.1074/jbc.M504716200
Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA (2004) Transcriptional regulatory code of a eukaryotic genome. Nature 431(7004):99–104. https://doi.org/10.1038/nature02800
Heer D, Sauer U (2008) Identification of furfural as a key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microb Biotechnol 1:497–506. https://doi.org/10.1111/j.1751-7915.2008.00050.x
Heer D, Heine D, Sauer U (2009) Resistance of Saccharomyces cerevisiae to high concentrations of furfural is based on NADPH-dependent reduction by at least two oxireductases. Appl Environ Microbiol 75(24):7631–7638. https://doi.org/10.1128/AEM.01649-09
Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425(6959):686–691. https://doi.org/10.1038/nature02026
Jayakody LN, Hayashi N, Kitagaki H (2011) Identification of glycolaldehyde as the key inhibitor of bioethanol fermentation by yeast and genome-wide analysis of its toxicity. Biotechnol Lett 33(2):285–292. https://doi.org/10.1007/s10529-010-0437-z
Jönsson LJ, Martín C (2016) Pretreatment of lignocellulose: formation of inhibitory by-products and strategies for minimizing their effects. Bioresour Technol 199:103–112. https://doi.org/10.1016/j.biortech.2015.10.009
Jörnvall H, Landreh M, Östberg LJ (2015) Alcohol dehydrogenase, SDR and MDR structural stages, present update and altered era. Chem Biol Interact 234:75–79. https://doi.org/10.1016/j.cbi.2014.10.017
Kallberg Y, Oppermann U, Jörnvall H, Persson B (2002) Short-chain dehydrogenases/reductases (SDRs): coenzyme-based functional assignments in completed genomes. Eur J Biochem 269(18):4409–4417. https://doi.org/10.1046/j.1432-1033.2002.03130.x
Kavanagh KL, Jörnvall H, Persson B, Oppermann U (2008) The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci 65:3895–3906. https://doi.org/10.1007/s00018-008-8588-y
Kawaguchi H, Hasunuma T, Ogino C, Kondo A (2016) Bioprocessing of bio-based chemicals produced from lignocellulosic feedstocks. Curr Opin Biotechnol 42:30–39. https://doi.org/10.1016/j.copbio.2016.02.031
Kumar AK, Sharma S (2017) Recent updates on different methods of pretreatment of lignocellulosic feedstocks: a review. Bioresour Bioprocess 4(1):7. https://doi.org/10.1186/s40643-017-0137-9
Laadan B, Almeida JR, Rådström P, Hahn-Hägerdal B, Gorwa-Grauslund M (2008) Identification of an NADH-dependent 5-hydroxymethylfurfural-reducing alcohol dehydrogenase in Saccharomyces cerevisiae. Yeast 25(3):191–198. https://doi.org/10.1002/yea.1578
Li X, Yang R, Ma M, Wang X, Tang J, Zhao X, Zhang X (2015) A novel aldehyde reductase encoded by YML131W from Saccharomyces cerevisiae confers tolerance to furfural derived from lignocellulosic biomass conversion. Bioenerg Res 8(1):119–129. https://doi.org/10.1007/s12155-014-9506-9
Lineweaver H, Burk D (1934) The determination of enzyme dissociation constants. J Am Chem Soc 56(3):658–666. https://doi.org/10.1021/ja01318a036
Liu ZL (2011) Molecular mechanisms of yeast tolerance and in situ detoxification of lignocellulose hydrolysates. Appl Microbiol Biotechnol 90:809–825. https://doi.org/10.1007/s00253-011-3167-9
Liu ZL, Moon J (2009) A novel NADPH-dependent aldehyde reductase gene from Saccharomyces cerevisiae NRRL Y-12632 involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Gene 446(1):1–10. https://doi.org/10.1016/j.gene.2009.06.018
Liu ZL, Slininger PJ (2007) Universal external RNA controls for microbial gene expression analysis using microarray and qRT-PCR. J Microbiol Methods 68(3):486–496. https://doi.org/10.1016/j.mimet.2006.10.014
Liu ZL, Moon J, Andersh BJ, Slininger PJ, Weber S (2008) Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 81:743–753. https://doi.org/10.1007/s00253-008-1702-0
Liu ZL, Palmquist DE, Ma M, Liu J, Alexander NJ (2009) Application of a master equation for quantitative mRNA analysis using qRT-PCR. J Biotechnol 143(1):10–16. https://doi.org/10.1016/j.jbiotec.2009.06.006
Ma M, Liu ZL (2010) Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4 and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genomics 11:660. https://doi.org/10.1186/1471-2164-11-660
Ma M, Wang X, Zhang X, Zhao X (2013) Alcohol dehydrogenases from Scheffersomyces stipitis involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Appl Microbiol Biotechnol 97(18):8411–8425. https://doi.org/10.1007/s00253-013-5110-8
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:D222–D226. https://doi.org/10.1093/nar/gku1221
Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. https://doi.org/10.1093/nar/gkw1129
Moon J, Liu ZL (2012) Engineered NADH-dependent GRE2 from Saccharomyces cerevisiae by directed enzyme evolution enhances HMF reduction using additional cofactor NADPH. Enzym Microb Technol 50(2):115–120. https://doi.org/10.1016/j.enzmictec.2011.10.007
Moon J, Liu ZL (2015) Direct enzyme assay evidence confirms aldehyde reductase function of Ydr541cp and Ygl039wp from Saccharomyces cerevisiae. Yeast 32(4):399–407. https://doi.org/10.1002/yea.3067
Persson B, Kallberg Y, Bray JE, Bruford E, Dellaporta SL, Favia AD, Duarte RG, Jörnvall H, Kavanagh KL, Kedishvili N, Kisiela M, Maser E, Mindnich R, Orchard S, Penning TM, Thornton JM, Adamski J, Oppermann U (2009) The SDR (short-chain dehydrogenase/reductase and related enzymes) nomenclature initiative. Chem Biol Interact 178(1–3):94–98. https://doi.org/10.1016/j.cbi.2008.10.040
Petersson A, Almeida JR, Modig T, Karhumaa K, Hahn-Hägerdal B, Gorwa-Grauslund MF, Lidén G (2006) A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast 23(6):455–464. https://doi.org/10.1002/yea.1370
Rabemanolontsoa H, Saka S (2016) Various pretreatments of lignocellulosics. Bioresour Technol 199:83–91. https://doi.org/10.1016/j.biortech.2015.08.029
Sgraja T, Ulschmid J, Becker K, Schneuwly S, Klebe G, Reuter K, Heine A (2004) Structural insights into the neuroprotective-acting carbonyl reductase sniffer of Drosophila melanogaster. J Mol Biol 342(5):1613–1624. https://doi.org/10.1016/j.jmb.2004.08.020
Sinicropi D, Cronin M, Liu ML (2007) Gene expression profiling utilizing microarray technology and RT-PCR. In: Ferrari M, Ozkan M, Heller M (eds) BioMEMS and biomedical nanotechnology, vol II: Micro/Nano technologies for genomics and proteomics. Springer-Verlag, Heidelberg, pp 23–46
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729. https://doi.org/10.1093/molbev/mst197
Teixeira MC, Monteiro PT, Guerreiro JF, Gonçalves JP, Mira NP, dos Santos SC, Cabrito TR, Palma M, Costa C, Francisco AP, Madeira SC, Oliveira AL, Freitas AT, Sá-Correia I (2014) The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucleic Acids Res 42:D161–D166. https://doi.org/10.1093/nar/gkt1015
Van Dijken JP, Scheffers WA (1986) Redox balances in the metabolism of sugars by yeasts. FEMS Microb Rev 32(3–4):199–224. https://doi.org/10.1016/0378-1097(86)90291-0
Wang X, Liang Z, Hou J, Bao X, Shen Y (2016) Identification and functional evaluation of the reductases and dehydrogenases from Saccharomyces cerevisiae involved in vanillin resistance. BMC Biotechnol 16:31. https://doi.org/10.1186/s12896-016-0264-y
Wang HY, Xiao DF, Zhou C, Wang LL, Wu L, YT L, Xiang QJ, Zhao K, Li X, Ma MG (2017) YLL056C from Saccharomyces cerevisiae encodes a novel protein with aldehyde reductase activity. Appl Microbiol Biotechnol 101(11):4507–4520. https://doi.org/10.1007/s00253-017-8209-5
Weber C, Farwick A, Benisch F, Brat D, Dietz H, Subtil T, Boles E (2010) Trends and challenges in the microbial production of lignocellulosic bioalcohol fuels. Appl Microbiol Biotechnol 87(4):1303–1315. https://doi.org/10.1007/s00253-010-2707-z
Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Véronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285(5429):901–906. https://doi.org/10.1126/science.285.5429.901
Yi X, Gu H, Gao Q, Liu ZL, Bao J (2015) Transcriptome analysis of Zymomonas mobilis ZM4 reveals mechanisms of tolerance and detoxification of phenolic aldehyde inhibitors from lignocellulose pretreatment. Biotechnol Biofuels 8:153. https://doi.org/10.1186/s13068-015-0333-9
Zhao X, Tang J, Wang X, Yang R, Zhang X, Gu Y, Li X, Ma M (2015) YNL134C from Saccharomyces cerevisiae encodes a novel protein with aldehyde reductase activity for detoxification of furfural derived from lignocellulosic biomass. Yeast 32(5):409–422. https://doi.org/10.1002/yea.3068
Acknowledgements
We are grateful to Dr. Z. Lewis Liu from Bioenergy Research Unit, NCAUR-ARS, U.S. Department of Agriculture (Peoria, IL, USA) for providing the reference mRNA control mix.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 31570086) and the Talent Introduction of Sichuan Agricultural University (No. 01426100).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals by any of the authors.
Electronic supplementary material
ESM 1
(PDF 215 kb)
Rights and permissions
About this article
Cite this article
Wang, H., Ouyang, Y., Zhou, C. et al. YKL071W from Saccharomyces cerevisiae encodes a novel aldehyde reductase for detoxification of glycolaldehyde and furfural derived from lignocellulose. Appl Microbiol Biotechnol 101, 8405–8418 (2017). https://doi.org/10.1007/s00253-017-8567-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8567-z