Abstract
Butyrophilins (BTN), specifically BTN3A, play a central role in the modulation of γδ T cells, which are mainly present in gut and mucosal tissues. BTN3A1 is known, for example, to activate Vγ9Vδ2 T cells by means of a phosphoantigen interaction. In the extended HLA region, three genes are located, designated BTN3A1, BTN3A2 and BTN3A3, which were also defined in rhesus macaques. In contrast to humans, rhesus monkeys have an additional gene, BTN3A3Like, which has the features of a pseudogene. cDNA analysis of 32 Indian rhesus and 16 cynomolgus macaques originating from multiple-generation families revealed that all three genes are oligomorphic, and the deduced amino acids display limited variation. The macaque BTN3A alleles segregated together with MHC alleles, proving their location in the extended (Major Histocompatibility Complex) MHC. BTN3A nearly full-length transcripts of macaques and humans cluster tightly together in the phylogenetic tree, suggesting that the genes represent true orthologs of each other. Despite the limited level of polymorphism, 15 Mamu- and 14 Mafa-BTN3A haplotypes were defined, and, as in humans, all three BTN3A genes are transcribed in PBMCs and colon tissues. In addition to regular full-length transcripts, a high number of various alternative splicing (AS) products were observed for all BTN3A alleles, which may result in different isoforms. The comparable function of certain subsets of γδ T cells in human and non-human primates in concert with high levels of sequence conservation observed for the BTN3A transcripts presents the opportunity to study these not yet well understood molecules in macaques as a model species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
T cells are an essential component of the adaptive immune system and use cell surface bound receptors (TCRs) to recognize ligands that may signal the presence of intracellular infections or cellular stress. In addition to T cells that express a T cell receptor composed of a heterodimer of an α and a β chain, another T cell lineage expresses a different set of receptors consisting of a γ and a δ chain instead. Although much less abundant than αβ T cells in peripheral blood, these γδ T cells are highly plentiful in tissues such as skin, the intestines and the reproductive tract (Adams et al. 2015). Moreover, they represent an important effector subset of innate-like T lymphocytes (Rhodes et al. 2016). In contrast to TCRs of the αβ lineage, however, which recognize antigenic peptides in the context of classical or non-classical MHC class I (MHC-I) molecules, some γδ-T cells recognize lipid molecules presented by MHC-like molecules such as CD1d and CD1c. Additionally, there are others, which can directly detect antigens, sometimes even without presentation via a canonical, antigen-presenting MHC or MHC-like molecule (Bai et al. 2012; Luoma et al. 2014; Roy et al. 2014). The most thoroughly investigated subset represents the Vγ9Vδ2 T cells, which are the dominant population of γδ-T cells in human peripheral blood, with potent activity regarding certain microbial infections and cancers (Gober et al. 2003; Morita et al. 2007). Vγ9Vδ2 T cells respond specifically to pyrophosphates derived from microbial sources or malignant cells. These phosphoantigens (pAg) are recognized in a TCR-dependent manner, but in contrast to αβ-T cells, they apparently do not require MHC molecules for pAg-dependent activation. Recently, molecules belonging to the butyrophilin (BTN) protein family, specifically butyrophilin-3A (BTN3A), were identified as an essential component in the pAg activation pathway of Vγ9Vδ2 T cells (Abeler-Dorner et al. 2012; Afrache et al. 2012; Karunakaran and Herrmann 2014; Rhodes et al. 2016; Vavassori et al. 2013). BTN3A proteins are cell-surface receptors expressed on a broad variety of cell types, including immune cells and some malignant cells such as ovarian cancer (Arnett and Viney 2014). They belong to the Ig-like superfamily and are evolutionarily and structurally related to the cluster of B7 costimulatory molecules CD80 and CD86 (Abeler-Dorner et al. 2012; Afrache et al. 2012; Rhodes et al. 2016). They exhibit two extracellular domains, IgV and IgC (Williams and Barclay 1988), and a transmembrane part. In contrast to the B7 molecule, however, most BTN molecules also contain a cytosolic B30.2 (PRYSPRY) domain, which is separated from the transmembrane domain by several very short exons, which are called heptads (Rhodes et al. 2001). BTN3A genes belong to a multigene family, which maps in humans to the extended MHC class I region and comprises three genes: BTN3A1, BTN3A2 and BTN3A3 (Horton et al. 2004; Rhodes et al. 2001, 2016). BTN3A1 and BTN3A3 contain the intracellular B30.2 domain, which is absent in BTN3A2. Although the prominent role of BTN3A1 in pAg-induced Vγ9Vδ2 T cell activation is well documented, the working mechanism is not yet fully understood, and the extracellular ligand for BTN3A1 molecules is still unknown. There is strong support for the intracellular sensing of pAg through the B30.2 domain of BTN3A1-mediating Vγ9Vδ2 T cell activation (Harly et al. 2012; Palakodeti et al. 2012; Peigne et al. 2017; Rhodes et al. 2015; Sandstrom et al. 2014). Additionally, crystallographic studies illustrated that BTN3A1 molecules may exist in two potential dimer conformations, and pAg-induced conformational changes of the intracellular domain of BTN3A1 might also involve dimer conversion (Gu et al. 2017, 2018; Palakodeti et al. 2012). While BTN3A1 is necessary for pAg-induced Vγ9Vδ2 T cell activation, this molecule alone may not be sufficient to conduct the activation process. Two cytoskeletal mediators have been identified, and named Periplakin and RhoB GTPase, which interact directly with BTN3A1, and may as such be involved in the activation process (Rhodes et al. 2015; Sebestyen et al. 2016). Additionally, recent work postulates that ABC transporters play a role in BTN3A-dependent γδ T cell activation by way of cytokine secretion (Rhodes et al. 2018). Furthermore, a co-expression and multimerization of BTN3A1 with its sister molecules BTN3A2 and BTN3A3 may be necessary for their function, comparable to a concerted action as documented for other BTN molecules (Di Marco Barros et al. 2016; Gu et al. 2018; Rhodes et al. 2015).
BTN3A proteins indeed play a role in T cell proliferation and cytokine production, which was manifested through the use of BTN3A-specific antibodies. One of the antibodies, 20.1, triggers Vγ9Vδ2 T cell activation, whereas a different antibody, 103.2, works as an antagonist and blocks pAg-induced T cell activation (Arnett and Viney 2014; Harly et al. 2012). The functional importance of BTN3A molecules was illustrated in a xenotransplantation model by the agonist antibody 20.1, which was able to enhance the Vγ9Vδ2 T cell-mediated killing of leukaemia cells (Benyamine et al. 2016). Moreover, the influence of BTN3A1 in stimulating anti-tumour effector Vγ9Vδ2 T cells in zolodronate-treated colorectal cancer cells was documented as well (Zocchi et al. 2017).
Mutations in BTN genes may influence disease outcome, since polymorphisms in BTN3A2 are associated with an increased susceptibility to developing type I diabetes (Viken et al. 2009). BTN3A genes are highly conserved in evolution and have been defined not only in higher primates but also in some other placental mammals such as the alpaca (Karunakaran et al. 2014). The three human BTN3A genes are the result of two successful rounds of duplications during the radiation of primates (Afrache et al. 2017). All three BTN3A genes seem to be present in hominoids (humans and great apes) and Old World monkeys (OWM). The concerted evolution of these genes resulted in a strong homogenization of the IgV domain in hominoids, where the sequences of BTN3A1 and BTN3A3 are replaced by BTN3A2. This homogenization of the IgV domain appears to be absent in OWM (Afrache et al. 2017).
Despite the increasing interest in BTN3A as immune modulators of Vγ9Vδ2 T cells, nothing is known so far about the possible levels of polymorphism and their distribution in OWM species such as macaques. Therefore, large families of rhesus and cynomolgus macaques were analysed for their BTN3A genes by cloning and Sanger sequencing.
Already in the beginning of the century, alternative splicing (AS) was observed in addition to constitutive splicing by Sanger sequencing specifically for human BTN3A transcripts, indicating that AS occurs far more frequently in these genes than in others (Rhodes et al. 2001). With more recent NGS techniques, AS has been documented in various genes of the immune system as well, and the different splicing mechanisms have been extensively described (Bruijnestijn et al. 2018; Keren et al. 2010; Voorter et al. 2016). In the present study, various isoforms of macaque BTN3A transcripts that were generated by AS have been documented, and the most frequent splice events have been characterized in detail.
Material and methods
Animals
We analysed DNA samples of 32 rhesus macaques of Indian origin belonging to two families of four generations and 16 cynomolgus macaques of various origins including different Indonesian islands as well as the Indonesian continent, belonging to two families of two generations, respectively. These animals belong to outbred breeding colonies that are housed at the BPRC, the Netherlands, and had been extensively characterized beforehand for their MHC class I and II haplotypes (Doxiadis et al. 2013; de Groot et al. 2014).
Genomic DNA (gDNA) of exon 2 of one rhesus macaque family (family sire C68; no. 19) was analysed, followed by a cDNA analysis of all animals for exon 2 to exon 9 of the BTN3A1 and BTN3A3 genes, exon 2 to exon 7 of the BTN3A2 gene and exon 2 of the pseudogene BTN3ALlike.
Genomic DNA isolation and BTN3A exon 2 gDNA sequencing
gDNA of the selected animals was extracted from EDTA blood samples or from immortalized B cell lines using a standard salting out procedure. Exon 2 amplification was performed as described using adapted primers originally published by Rhodes and co-workers (Rhodes et al. 2001) (Suppl. Table 1). Amplicons were sequenced and analysed as described below.
RNA isolation, cDNA preparation, cloning and sequencing
RNA was isolated from immortalized B cell lines or freshly prepared PBMCs (RNeasy kit, Qiagen) and was used for the synthesis of cDNA with the RevertAid First Strand cDNA synthesis Kit (Thermo scientific). BTN3A1, BTN3A2 and BTN3A3 sequences (exon 1 [partly] to exon 9) were gained by PCR on cDNA using newly designed primer sets for the specific amplification of all three BTN3As. The amplification of BTN3A3Like alleles was only successful by using a specific exon 2 5′primer and an exon 4 3′primer (Suppl. Table 1). Primers were synthesized by Invitrogen. PCR was performed in a 50-μl reaction containing 0.02 U/μl of Phusion High-Fidelity DNA polymerase enzyme with 0.3 μM of each primer, 250 μM dNTPs, 1× Phusion buffer HF, 3%DMSO (Invitrogen, Life Technologies) and 200 ng cDNA. The cycling parameters were a 2-min 98 °C initial denaturation step, followed by 30 cycles (35 cycles for BTN3A1) of a 30-s 98 °C denaturation step, a 30-s annealing step (54 °C for BTN3A1, 59 °C for BTN3A2 and 55 °C for BTN3A3) and a 90-s 72 °C extension step. A final extension step was performed at 72 °C for 7 min. PCR products were purified using a geneJet Gel Extraction Kit (Thermo Scientific™), and the purified amplicons were cloned into the pJET vector using the CloneJET PCR cloning kit, both according to the manufacturer’s guidelines (Thermo Scientific™). The cloned amplicons were then transformed in Escherichia coli-XL1-blue cells with the TransformAid Bacterial Transformation Kit (Thermo Scientific™). Per animal, a minimum of 32 clones was selected, and plasmid DNA was isolated using a standard mini-preparation procedure. The purified plasmid DNA was sequenced on an ABI 3500 genetic analyser (Applied Biosystems, Foster City, USA). The sequencing reaction was performed by 2 μM pJET primer, 1 μl BigDye terminator and 2 μl of 5× sequencing buffer in a total volume of 10 μl (Thermo Scientific™). The resulting sequences were analysed using the Sequence Navigator program (Applied Biosystems, Foster City, USA). BTN sequences were revised manually by applying the Lasergene 12 SeqMan Pro Sequence Alignment Editor. Allele definitions are based on at least two clones with identical sequences from different monkeys or on independent PCRs from one monkey. The sequences were named in accordance with the general nomenclature rules of the ImmunoPolymorphismDatabase (IPD-MHC database of nonhuman primates [NHP]) (de Groot et al. 2012) and sent to the NCBI database (Accession numbers: BTN3A1: LT992538-LT992545 and LT992494-LT992501; BTN3A2: LT992546-LT992555; BTN3A3: LT993304-LT993321 and BTN3A1L: LT993431-LT993437). The BTN3A3 pseudogene sequence of the olive baboon was extracted from the NCBI database (Accession no. XM_009198763).
Phylogenetic analysis of exon 2 and of exons 2–6 of BTN3A alleles
The evolutionary history of the BTNA3 alleles was inferred using the neighbour-joining method and the optimal tree. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method and are in the units of the number of base substitutions per site. The analyses involved 37 (gDNA exon 2) and 56 (cDNA exon 2 to exon 6) sequences, respectively. Codon positions included were 1st + 2nd + 3rd + noncoding. All ambiguous positions were removed for each sequence pair. There was a total of 348 (exon 2) and 879 (exon 2 to exon 6) positions, respectively, in the final dataset. Evolutionary analyses were conducted in MEGA7.
Alternative splicing
Alternative splice products observed for all BTN3A transcripts were categorized according to their frequency and splice mechanism (Suppl. Table 2). All observed splice events were substantiated by the prediction and scoring of the alternative and actual splice sites using three different in silico prediction tools. The selected tools were the Maximum Entropy Modeling Scan (MaxEntScan; MES) (Yeo and Burge 2004), the Position Weight Matrix (PWM) via SpliceView (Rogozin and Milanesi 1997) and the Human Splice Finder (HSF) (Desmet et al. 2009). These tools use different output value ranges, but the higher score always implies a better-predicted splice site. It should be noted that whether or not a splice event occurs, the scores do not provide a threshold, and the scores should only be used to compare related splice sites.
Results
Exon 2 analyses of Mamu-BTN3A genes
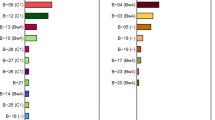
To scan for polymorphism of BTN3A genes in macaques, exon 2, encoding the IgV domain of the BTN3A genes of a four-generation, MHC-typed rhesus macaque family (family sire C68; #19), was sequenced at the genomic as well as the transcription level. All three BTN3A genes (BTN3A1, 3A2 and 3A3) are present in the rhesus macaque. BTN3A1 and BTN3A3 are oligomorphic with four and three alleles, respectively, whereas in the studied cohort, BTN3A2 did not show allelic variation in exon 2. Additionally, a fourth BTN3A gene was identified, seven alleles of which have been differentiated. Based on the similarity of this gene with BTN3A3 sequences of other Old World Monkeys (OWM) extracted from the NCBI database, it was provisionally named BTN3A3Like. In rhesus and cynomolgus macaques, the sequences of this gene share a stop codon at triplet 65 of exon 2 near the end of this exon, leading to a truncated gene product, and therefore, this gene appears to be a pseudogene. Phylogenetic analyses of the rhesus macaque exon 2 alleles of the BTN3A1, BTN3A2 and BTN3A3 genes showed that they cluster together with alleles of the same gene of other OWM. In contrast, hominoid (Hom) exon 2 sequences of the three BTN3A genes fall apart from the gene clusters of OWM and do not branch in a gene-specific manner. Macaque BTN3A3Like alleles, however, constitute a separate cluster with a long branch length (Fig. 1), suggesting that this gene has undergone many mutations during evolution.
Phylogenetic tree of exon 2 of BTN3A1, BTN3A2, BTN3A3 and BTN3A3Like of diverse primate species. Exon 2 sequences of the BTN3A genes and the BTN3ALike pseudogene of the rhesus macaque family EAW have been compared to BTN3A exon 2 sequences of humans (Hosa) and to various non-human primate species as deposited in the NCBI database. Gogo Gorilla gorilla, Patr Pan troglodytes, Papa Pan paniscus, Paan Papio anubis, Mamu Macaca mulatta, Mafa Macaca fascicularis, Mane Macaca nemestrina, Male Mandrillus leucophaeus
Mamu- and Mafa-BTN3A allele discovery
To determine the degree of allelic polymorphism in other exons and to assess whether the alleles defined in exon 2 represent bonafide transcripts, we analysed cDNA from exons 2–7 (partly) of BTN3A2 and exons 2–9 of BTN3A1/BTN3A3. For BTN3A3Like, only exons 2 and 3 could be successfully amplified. However, like in exon 2, exon 3 of all BTN3A3Like alleles showed various stop codons, and therefore, no functional BTN products may be expected from these transcripts. This is why we did not use these sequences for further analyses as sequence or deduced amino acid comparisons. The analyses of the BTN3A transcripts of BTN3A1, BTN3A2 and BTN3A3 were performed for animals from the previously mentioned rhesus macaque family (sire C68, #19) (Fig. 2) and an additional four-generation rhesus macaque family (sire EAW; #13; Suppl. Fig. 1). The corresponding transcripts of all BTN3A alleles, which had been defined beforehand for exon 2, could be recovered. Most of them are shared between the two rhesus macaque families, and a total of eight BTN3A1 and six BTN3A3 alleles could be differentiated, which gave rise to seven and five different allotypes, respectively (Fig. 3a, b). With regard to polymorphic sites, only a few amino acid replacements were observed within the BTN3A1 and BTN3A3 genes of rhesus macaques, which suggest a high level of conservation. BTN3A2 has a stop codon in exon 7, and therefore, as in humans, no B30.2 intracellular part is present at the transcript level. This molecule shows a very low degree of variation, with only three allotypes (Fig. 3c).
Pedigree of rhesus macaque family C68 and their deduced BTN3A haplotypes. Blue squares represent male and red ovals female animals. Grey squares/ovals indicate animals that have not been typed for BTN3A. Below the pedigree, the BTN3A haplotypes (designated a-o) are listed and the number of appearance (number sign) in family 1 (C68) and/or family 2 (EAW; Suppl. Fig. 1). For each animal, the inherited BTN3A haplotype is indicated in the pedigrees
In addition, 16 cynomolgus macaques belonging to two families were analysed for their BTN3A polymorphism. Although fewer animals were tested than for rhesus macaques, an identical number of BTN3A1 alleles have been recorded in both the cynomolgus and the rhesus macaque cohort, namely, eight of them, which also gave rise to seven different allotypes. Accordingly, cynomolgus macaques show more allelic variation for BTN3A2 and BTN3A3 with six and 12 alleles, which result in five and six different allotypes, respectively (Fig. 3a–c). In contrast to the rhesus macaque, in which both families share most of the BTN3A alleles, the two cynomolgus macaque families show mainly different alleles for BTN3A1 and BTN3A3 (Fig. 4).
Pedigree of two cynomolgus macaque families and their deduced BTN3A haplotypes. Blue squares represent male, and red ovals represent female animals. Below the pedigree, the deduced BTN3A haplotypes (designated a–n) are listed, and the appearance in family 1 or 2 is indicated. For each animal, the inherited haplotype is indicated in the pedigrees
Exons 7–9 of BTN3A genes have been excluded for phylogenetic analysis, since they are not comparable for all three BTN3A genes. The analysis of exons 2–6 showed that the alleles defined in this study cluster together with alleles of the same BTN3A genes of all other species analysed including hominoids (Fig. 5). This result confirms the observation that macaque BTN3A genes are true orthologs of their human equivalents (Afrache et al. 2017). In addition, the short branch lengths in the phylogenetic tree indicate that variation of the BTN3A genes is very low with regard to the number of variable nucleotides per allele. BTN3A2 is the most conserved entity, as reflected by the lowest nucleotide and amino acid variation (Fig. 3a–c).
Phylogenetic tree of exons 2–6 of BTN3A1, BTN3A2 and BTN3A3 of different primate species. Sequences of the BTN3A genes defined in the two rhesus macaque and the two cynomolgus families have been compared to BTN3A sequences of humans (Hosa) and various NHP species as deposited in the NCBI database. Gogo Gorilla gorilla, Patr Pan troglodytes, Mamu Macaca mulatta, Mafa Macaca fascicularis, Mane Macaca nemestrina
Furthermore, the positions of the variable, macaque-specific amino acids of all three BTN3A gene products are scattered over the coding sequence and not concentrated on certain positions (Fig. 3a–c), as contrasts the situation for the peptide binding site of the polymorphic MHC molecules (Hughes and Nei 1988). Exon 2 of BTN3A1, however, which encodes the IgV domain, forms an exception; the same four variable, macaque-specific amino acid positions are encoded within this exon, resulting in two different amino acids (Fig. 3a).
Mamu- and Mafa-BTN3A haplotype definition
Mapping of the various Mamu- and Mafa-BTN genes against published rhesus and cynomolgus whole genome sequences, respectively, showed the positions of the different Mamu- and Mafa-BTN1/2/3A genes on the extended MHC (https://www.ensembl.org/Macaca_mulatta/Info;https://www.ensembl.org/Macaca_fascicularis/Info/), which are highly comparable to the positions of the human BTN genes (Horton et al. 2004). (Fig. 6).
Schematic illustration of the localization of the BTN1/2/3A genes of humans, rhesus and cynomolgus macaques on the extended MHC class I region. The pseudogenes Hosa-BTN2A3ps and Mamu/Mafa-BTN3A3ps are marked by dark green squares. Although clearly defined in our experiments, Mafa-BTN3A3 was not detected on the whole genome sequence (NC_022275.1). Therefore, its possible localization is indicated by an unfilled square. aHorton et al. (2004)
The co-segregation of the alleles of all three BTN3A genes and those of the BTN3A3Like pseudogene within the macaque families confirm duplication of the BTN3A genes and their localization in close proximity to each other on the chromosome. We were able to define 15 and 14 BTN3A haplotypes in the two rhesus and cynomolgus macaque families, respectively. Figure 2 and Suppl. Fig. 1 show the pedigrees of the two rhesus macaque families with their deduced BTN3A haplotypes, whereas in Fig. 4, the pedigrees of both cynomolgus macaque families and deduced BTN3A haplotypes are illustrated. Whereas the two rhesus macaque families share about half of their haplotypes, the two cynomolgus macaque families have no BTN3A haplotypes in common.
Additionally, mapping of the BTN3A genes within the extended MHC complex of the rhesus macaque is confirmed by the co-segregation of most of the BTN3A haplotypes with the MHC haplotypes defined beforehand. The co-segregation of MHC haplotypes and BTN3A alleles is also observed in the two cynomolgus macaque families (data not shown). As expected in accordance with the long distance between both regions (> 3 Mb), crossing over events between BTN3A and MHC haplotypes can be postulated as well as point mutations, leading to new allele/haplotype definitions. The deduced MHC-BTN3A haplotypes of the rhesus macaque family 2 are given as examples (Suppl. Fig. 1).
AS of BTN3A in macaque species
For many gene systems, high numbers of AS products showed up by way of the introduction of long-range next-generation sequencing (NGS) as PacBio Single Molecule Real-Time (SMART) sequencing compared to Sanger sequencing. By using Sanger sequencing of the different BTN3A cDNAs, however, an unexpectedly high number of various AS transcripts were observed in both macaque species in addition to the constitutive transcripts. AS products of BTN3A of nine rhesus macaques belonging to family 1 (Fig. 2) were analysed in detail. AS events, which have been observed more than three times, were categorized according to their splicing events as reviewed by Ast (2004) and Keren and co-workers (Keren et al. 2010). Figure 7 provides a summary of the different AS events, which may generate a shorter but in-frame transcript. We have observed exon skipping not only of exons 3, 4, 5 and 7 but also of multiple exons together (exons 3 + 4 and exons 3 to 5). Additionally, splice events mediated by alternative splice sites (SS), alternative 3′ SS and alternative 5′ SS were frequently observed, whereas in-frame splice events mediated by intron retention and cryptic exon inclusion were not noted. The alternatively spliced transcripts potentially result in eight different isoforms (Fig. 7b (a–h)): There are molecules that have an IgV and an IgC domain but no intracellular B30.2 part (Fig. 7b (a)); molecules with IgV, IgC and a B30.2 part but a heptad missing (Fig. 7b (b)); and molecules with only the IgC (Fig. 7b (c)) or the IgV (Fig. 7b (d, e)) domain. Additionally, AS transcripts are observed without the exons encoding the transmembrane part (Fig. 7b (f–h)), which could therefore result in soluble BTN3A isoforms. None of the observed splice events resulted in a soluble IgC domain. Exon skipping is the most frequently observed splicing mechanism that resulted in in-frame transcripts, as previously described for eukaryotes in general (Keren et al. 2010). Of all exon-skipping events, exon 4 skipping is the most frequent in BTN3A1 (#18) and BTN3A2 (#17) but has not been detected for BTN3A3. To substantiate the observed splice event and its absence in BTN3A3, we predicted and scored the alternative splice sites and compared them with each other using three different prediction models: namely, MaxEntScan (MES), Human Splice Finder (HSF) and Position Weight Matrix (PWM). These models did substantiate the observed skipping of exon 4 in transcripts of BTN3A1 and BTN3A2, but the splicing strength scores could not explain the absence of this splice event in BTN3A3 transcripts (Suppl. Table 2).
Schematic illustration of the BTN3A gene sequences, their resulting transcripts and proteins. aBTN3A1, BTN3A3 and BTN3A2 genes, transcripts and predicted proteins using actual splice sites. b Various alternative splicing (AS) transcripts of BTN3A genes as observed in rhesus macaque family C68. Transcripts are categorized by the AS mechanism. These AS transcripts do not possess a premature stop codon and should therefore lead to a shorter but intact protein. c Illustration of one specific AS event occurring in all BTN3A genes and the pseudogene transcripts. This AS involves alternative 3′ splice site (SS) and 5′ SS selection within exon 2, which creates a 112-bp deletion in the middle of exon 2 and a frame shift (indicated in green) leading to a stop codon at the end of exon 2 (asterisk). Therefore, no gene product is expected
Nearly as often, splice events mediated by alternative 3′ SS (#6, #4, #10) were detected, whereas splice events mediated by alternative 5′ SS (#5) were far less frequently observed (Fig. 7b). Additionally, alternative splice sites resulted in the partial retention of introns, often in combination with exon skipping, but always introducing a stop codon so that no intact gene product would be formed. One of these splice events resulted in a deletion of 112 bp, and was mediated by alternative 3′ and 5′ splice sites in exon 2. This AS was frequently observed in all three BTN3A transcripts and the BTN3ALike pseudogene (Fig. 7c), although no bona fide gene product could be expected. All three in silico models predicted identical scores for the alternative splice sites that mediate the event in the transcripts of BTN3A (pseudo)genes. The PWM model did not detect the alternative 3′ SS at all. Compared to the actual splice site scores, the alternative splice site scores were lower for all models, suggesting that constitutive splicing is dominant over this alternative splicing event (Suppl. Table 2).
Discussion
The distribution analysis of BTN3A genes in rhesus and cynomolgus macaque families confirmed the presence of the three genes BTN3A1, BTN3A2 and BTN3A3 in all animals tested (Afrache et al. 2017). In addition, the discovery of a pseudogene, named BTN3A3Like, was facilitated by exon 2 analysis. This pseudogene appears, however, to be absent in hominoids, who instead have a BTN2A pseudogene (Rhodes et al. 2016). This observation confirms the flexibility of the region not only in humans but also in OWM such as macaques.
Allelic variation of all BTN3A genes was documented for rhesus as well as cynomolgus macaques, although the degree of polymorphism is quite low, with the lowest variation observed for BTN3A2 followed by BTN3A3 and BTN3A1. The latter gene has, however, a remarkably conserved exon 2 (IgV region), and no non-synonymous mutations in exon 4 (TM) and exons 6–8 (heptads) were observed. BTN3A3 shows no non-synonymous variations in exons 5–8 (heptads) and only one in exon 4 (TM region) and exon 9 (intracellular B30.2 part), respectively. As may be expected for a pseudogene, which experiences no purifying selection, the allelic variation observed in exon 2 of BTN3A3Like is higher than in those of the BTN3A genes (Fig. 2). In concordance with the level of polymorphism observed in macaque MHC genes (Doxiadis et al. 2013), BTN3A alleles and haplotypes are more variable in cynomolgus monkeys than in rhesus macaques of Indian origin as represented in our cohort.
Furthermore, phylogenetic analysis of exon 2 showed that HOM exon 2 sequences of the three BTN3A genes fall apart from the gene clusters of OWM and that they do not branch in a gene-specific manner as do the alleles of OWM (Fig. 1). The homogenization, replacing IgV-encoding sequences of BTN3A1, BTN3A2 and BTN3A3 in HOM, has recently been described (Afrache et al. 2017). This phenomenon appears to be the result of an ancient recombination between the 5′ region of a BTN3A1Like gene and the 3′ region of a BTN3A2-like gene, resulting in a chimeric BTN3A3 gene. Our data suggest that this ancient recombination process took place after the divergence of OWM and HOM ~ 25 myr ago, since the homogenization is not observed in OWM.
In addition to the exon 2 sequences of BTN3A3Like genes of diverse macaque species (Fig. 1), a database search revealed the presence of a BTN3A3Like sequence in the olive baboon. In contrast to the macaque pseudogene sequences, however, which all have the stop codon at amino acid 94 in exon 2, the olive baboon sequence already has a stop codon at amino acid 32. In the latter, a sequence of 258 bp is integrated after exon 1, which seems to originate from the 3′ untranslated region of the BTN3A2 gene. The existence of such a chimeric pseudogene seems to provide additional evidence that intragenic recombination processes are a common feature within the BTN region.
In contrast to the phylogenetic analysis of exon 2, the phylogeny of the BTN3A transcripts of exons 2–6 of human and non-human primate sequences shows a gene-specific clustering (Fig. 5), confirming that macaque BTN3A genes are true orthologs of their human equivalents.
Based on the relationship of the animals and segregation profiles within the rhesus and cynomolgus macaque families, we were able to define several BTN3A haplotypes. In concordance with the higher polymorphism observed in cynomolgus monkeys, haplotypes are more variable in this species than they are in rhesus macaques, most probably due to their various origins from different parts of Indonesia. Physical mapping showed the localization of the different macaque BTN genes within the extended MHC class I region, which is comparable to the position of the human BTN region (Fig. 6). Frequent co-segregation of BTN3A alleles/haplotypes with the MHC haplotypes verifies their localization as has been described for humans (Horton et al. 2004).
Already at the beginning of the century, AS was observed by Sanger sequencing for human BTN3A transcripts, indicating that AS occurs far more frequently in these genes than in others (Rhodes et al. 2001). Recently, in BG genes, which represent family members of the BTN gene family in chicken, AS has also been defined by Sanger sequencing (Chen et al. 2018). A remarkable phenomenon is the high degree of AS that has been observed for alleles of all BTN3A genes and the pseudogene. In general—e.g. for MHC class I and II genes—AS is rarely revealed using the low-resolution Sanger sequencing technique and is only detected when NGS techniques have been used and even then in relatively low numbers. The fact that AS isoforms are detected by the Sanger method for macaque BTN3A genes suggests that these transcripts are generated at a fairly high frequency. One could imagine that isoforms of the BTN3A molecules that have been generated due to AS may play a specific role as Vγ9Vδ2 T cell modulators. Exon skipping comprised the most frequently observed AS events, especially those that included exon 4, which encodes for the transmembrane part section, as was observed for BTN3A1 and BTN3A2. Predicting the actual splice sites with three different in silico models did not answer the question as to why exon 4 skipping does not occur in BTN3A3. Intron 3, which is situated next to exon 4, does show a high degree of nucleotide variation between the different BTN3A genes. Moreover, intron 3 of BTN3A3 is longer than the equivalent structure of the other two BTN3A genes. All these differences may influence alternative splicing—for example, through the existence of additional silencer/enhancer motives and/or the formation of secondary structures, which may prevent the appearance of a certain exon within the transcript. It is therefore plausible to suggest that the intron sequences may be the reason that this AS event can be detected in the transcripts of only two of the three BTN3A genes.
Exon skipping that includes exon 4, which encodes the transmembrane section, may result in the generation of soluble molecules. Gu et al. (2018) have presented a model that postulates that BTN3A isoforms help BTN3A1, the pAg-sensing molecule expressed on the cell surface, to make a conformational change when activated. This change, including possible multimerization events, can then be recognized by Vγ9Vδ2 T cell receptors. Soluble molecules may be especially supportive in this process. In this case, the frequently observed AS isoforms may actually represent a regulation phenomenon that helps in activating BTN3A-positive cells. We mainly looked at transcripts of white blood cells, which are a proxy for what may happen elsewhere in the body. One wonders of course whether the AS profiles would be different in tissues in healthy or stressed conditions. Indeed, soluble BTN3A isoforms are detected in the supernatant of pancreatic cell lines and the plasma of pancreatic ductal adenocarcinoma patients. Soluble BTN3A and BTN3A1 were associated with a decreased overall survival rate of these patients (Benyamine et al. 2016; Blazquez et al. 2018). Consequently, soluble BTN3A molecules can serve as prognostic biomarkers, highlighting the value of macaques in clinical cancer research, in addition to their importance in future proof-of-concept studies for the protective features of Vγ9Vδ2 T cells—for instance, against M. tuberculosis or other infectious agents (Chen 2016).
References
Abeler-Dorner L, Swamy M, Williams G, Hayday AC, Bas A (2012) Butyrophilins: an emerging family of immune regulators. Trends Immunol 33:34–41
Adams EJ, Gu S, Luoma AM (2015) Human gamma delta T cells: evolution and ligand recognition. Cell Immunol 296:31–40
Afrache H, Gouret P, Ainouche S, Pontarotti P, Olive D (2012) The butyrophilin (BTN) gene family: from milk fat to the regulation of the immune response. Immunogenetics 64:781–794
Afrache H, Pontarotti P, Abi-Rached L, Olive D (2017) Evolutionary and polymorphism analyses reveal the central role of BTN3A2 in the concerted evolution of the BTN3 gene family. Immunogenetics 69:379–390
Arnett HA, Viney JL (2014) Immune modulation by butyrophilins. Nat Rev Immunol 14:559–569
Ast G (2004) How did alternative splicing evolve? Nat Rev Genet 5:773–782
Bai L, Picard D, Anderson B, Chaudhary V, Luoma A, Jabri B, Adams EJ, Savage PB, Bendelac A (2012) The majority of CD1d-sulfatide-specific T cells in human blood use a semiinvariant Vdelta1 TCR. Eur J Immunol 42:2505–2510
Benyamine A, Le Roy A, Mamessier E, Gertner-Dardenne J, Castanier C, Orlanducci F, Pouyet L, Goubard A, Collette Y, Vey N, Scotet E, Castellano R, Olive D (2016) BTN3A molecules considerably improve Vgamma9Vdelta2T cells-based immunotherapy in acute myeloid leukemia. Oncoimmunology 5:e1146843
Blazquez JL, Benyamine A, Pasero C, Olive D (2018) New insights into the regulation of gammadelta T cells by BTN3A and other BTN/BTNL in tumor immunity. Front Immunol 9:1601
Chen ZW (2016) Protective immune responses of major Vgamma2Vdelta2 T-cell subset in M. tuberculosis infection. Curr Opin Immunol 42:105–112
Chen L, Fakiola M, Staines K, Butter C, Kaufman J (2018) Functional alleles of chicken BG genes, members of the butyrophilin gene family, in peripheral T cells. Front Immunol 9:930
de Groot NG, Otting N, Robinson J, Blancher A, Lafont BA, Marsh S, O’Connor DH, Shiina T, Walter L, Watkins DI, Bontrop RE (2012) Nomenclature report on the major histocompatibility complex genes and alleles of Great Apes, Old and New World monkey species. Immunogenetics 64:615–631
de Groot N, Doxiadis GG, Otting N, de Vos-Rouweler AJ, Bontrop RE (2014) Differential recombination dynamics within the MHC of macaque species. Immunogenetics 66:535–544
Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37:e67
Di Marco Barros R, Roberts NA, Dart RJ, Vantourout P, Jandke A, Nussbaumer O, Deban L, Cipolat S, Hart R, Iannitto ML, Laing A, Spencer-Dene B, East P, Gibbons D, Irving PM, Pereira P, Steinhoff U, Hayday A (2016) Epithelia use butyrophilin-like molecules to shape organ-specific gammadelta T cell compartments. Cell 167:203–218 e217
Doxiadis GG, de Groot N, Otting N, de Vos-Rouweler AJ, Bolijn MJ, Heijmans CM, de Groot NG, van der Wiel MK, Remarque EJ, Vangenot C, Nunes JM, Sanchez-Mazas A, Bontrop RE (2013) Haplotype diversity generated by ancient recombination-like events in the MHC of Indian rhesus macaques. Immunogenetics 65:569–584
Gober HJ, Kistowska M, Angman L, Jeno P, Mori L, De Libero G (2003) Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J Exp Med 197:163–168
Gu S, Sachleben JR, Boughter CT, Nawrocka WI, Borowska MT, Tarrasch JT, Skiniotis G, Roux B, Adams EJ (2017) Phosphoantigen-induced conformational change of butyrophilin 3A1 (BTN3A1) and its implication on Vgamma9Vdelta2 T cell activation. Proc Natl Acad Sci U S A 114:E7311–E7320
Gu S, Borowska MT, Boughter CT, Adams EJ (2018) Butyrophilin3A proteins and Vgamma9Vdelta2 T cell activation. Semin Cell Dev Biol 84:65–74
Harly C, Guillaume Y, Nedellec S, Peigne CM, Monkkonen H, Monkkonen J, Li J, Kuball J, Adams EJ, Netzer S, Dechanet-Merville J, Leger A, Herrmann T, Breathnach R, Olive D, Bonneville M, Scotet E (2012) Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human gammadelta T-cell subset. Blood 120:2269–2279
Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC Jr, Wright MW, Wain HM, Trowsdale J, Ziegler A, Beck S (2004) Gene map of the extended human MHC. Nat Rev Genet 5:889–899
Hughes AL, Nei M (1988) Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 355:167–170
Jesse Bruijnesteijn, Marit KH, van der Wiel, Nanine de Groot, Nel Otting, Annemiek JM, de Vos-Rouweler, Neubury M. Lardy, Natasja G. de Groot, Ronald E. Bontrop, (2018) Extensive Alternative Splicing of KIR Transcripts. Frontiers in Immunology 9
Karunakaran MM, Herrmann T (2014) The Vgamma9Vdelta2 T cell antigen receptor and butyrophilin-3 A1: models of interaction, the possibility of co-evolution, and the case of dendritic epidermal T cells. Front Immunol 5:648
Karunakaran MM, Gobel TW, Starick L, Walter L, Herrmann T (2014) Vgamma9 and Vdelta2 T cell antigen receptor genes and butyrophilin 3 (BTN3) emerged with placental mammals and are concomitantly preserved in selected species like alpaca (Vicugna pacos). Immunogenetics 66:243–254
Keren H, Lev-Maor G, Ast G (2010) Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11:345–355
Luoma AM, Castro CD, Adams EJ (2014) gammadelta T cell surveillance via CD1 molecules. Trends Immunol 35:613–621
Morita CT, Jin C, Sarikonda G, Wang H (2007) Nonpeptide antigens, presentation mechanisms, and immunological memory of human Vgamma2Vdelta2 T cells: discriminating friend from foe through the recognition of prenyl pyrophosphate antigens. Immunol Rev 215:59–76
Palakodeti A, Sandstrom A, Sundaresan L, Harly C, Nedellec S, Olive D, Scotet E, Bonneville M, Adams EJ (2012) The molecular basis for modulation of human Vgamma9Vdelta2 T cell responses by CD277/butyrophilin-3 (BTN3A)-specific antibodies. J Biol Chem 287:32780–32790
Peigne CM, Leger A, Gesnel MC, Konczak F, Olive D, Bonneville M, Breathnach R, Scotet E (2017) The juxtamembrane domain of butyrophilin BTN3A1 controls phosphoantigen-mediated activation of human Vgamma9Vdelta2 T cells. J Immunol 198:4228–4234
Rhodes DA, Stammers M, Malcherek G, Beck S, Trowsdale J (2001) The cluster of BTN genes in the extended major histocompatibility complex. Genomics 71:351–362
Rhodes DA, Chen HC, Price AJ, Keeble AH, Davey MS, James LC, Eberl M, Trowsdale J (2015) Activation of human gammadelta T cells by cytosolic interactions of BTN3A1 with soluble phosphoantigens and the cytoskeletal adaptor periplakin. J Immunol 194:2390–2398
Rhodes DA, Reith W, Trowsdale J (2016) Regulation of immunity by butyrophilins. Annu Rev Immunol 34:151–172
Rhodes DA, Chen HC, Williamson JC, Hill A, Yuan J, Smith S, Rhodes H, Trowsdale J, Lehner PJ, Herrmann T, Eberl M (2018) Regulation of human gammadelta T cells by BTN3A1 protein stability and ATP-binding cassette transporters. Front Immunol 9:662
Rogozin IB, Milanesi L (1997) Analysis of donor splice sites in different eukaryotic organisms. J Mol Evol 45:50–59
Roy S, Ly D, Li NS, Altman JD, Piccirilli JA, Moody DB, Adams EJ (2014) Molecular basis of mycobacterial lipid antigen presentation by CD1c and its recognition by alphabeta T cells. Proc Natl Acad Sci U S A 111:E4648–E4657
Sandstrom A, Peigne CM, Leger A, Crooks JE, Konczak F, Gesnel MC, Breathnach R, Bonneville M, Scotet E, Adams EJ (2014) The intracellular B30.2 domain of butyrophilin 3A1 binds phosphoantigens to mediate activation of human Vgamma9Vdelta2 T cells. Immunity 40:490–500
Sebestyen Z, Scheper W, Vyborova A, Gu S, Rychnavska Z, Schiffler M, Cleven A, Cheneau C, van Noorden M, Peigne CM, Olive D, Lebbink RJ, Oostvogels R, Mutis T, Schuurhuis GJ, Adams EJ, Scotet E, Kuball J (2016) RhoB mediates phosphoantigen recognition by Vgamma9Vdelta2 T cell receptor. Cell Rep 15:1973–1985
Vavassori S, Kumar A, Wan GS, Ramanjaneyulu GS, Cavallari M, El Daker S, Beddoe T, Theodossis A, Williams NK, Gostick E, Price DA, Soudamini DU, Voon KK, Olivo M, Rossjohn J, Mori L, De Libero G (2013) Butyrophilin 3A1 binds phosphorylated antigens and stimulates human gammadelta T cells. Nat Immunol 14:908–916
Viken MK, Blonhoff A, Olsson M, Akselsen HE, Pociot F, Nerup J, Kockum I, Cambon-Thomsen A, Thorsby E, Undlien DE, Lie BA (2009) Reproducible association with type 1 diabetes in the extended class I region of the major histocompatibility complex. Genes Immun 10:323–333
Voorter CEM, Gerritsen KEH, Groeneweg M, Wieten L, Tilanus MGJ (2016) The role of gene polymorphism in HLA class I splicing. International Journal of Immunogenetics 43 (2):65–78
Williams AF, Barclay AN (1988) The immunoglobulin superfamily--domains for cell surface recognition. Annu Rev Immunol 6:381–405
Yeo G, Burge CB (2004) Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11:377–394
Zocchi MR, Costa D, Vene R, Tosetti F, Ferrari N, Minghelli S, Benelli R, Scabini S, Romairone E, Catellani S, Profumo A, Poggi A (2017) Zoledronate can induce colorectal cancer microenvironment expressing BTN3A1 to stimulate effector gammadelta T cells with antitumor activity. Oncoimmunology 6:e1278099
Acknowledgements
The authors wish to thank F. van Hassel for preparing the figures and D. Devine for editing the manuscript.
Funding
This work was supported in part by NIAID contracts HHSN272201100013C and HHSN272201600007C.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All applicable international, national and/or institutional guidelines for the care and use of animals were followed.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Suppl. Fig. 1.
Pedigree of rhesus macaque family EAW with deduced BTN3A and MHC haplotypes. Blue squares represent male and red ovals female animals. Grey squares/ovals indicate the animals that have not been typed for BTN3A. Letters (a-o) indicate the BTN3A haplotype defined in this family. Below the pedigree, the BTN3A haplotypes (a-o) are listed. The subdivision in a1 or a2 and in c1-c4 indicates that a certain BTN3A haplotype is associated with a different MHC haplotype. (PDF 333 kb)
Suppl. Table 1
(DOCX 18 kb)
Suppl. Table 2
(XLSX 11 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
de Groot, N., Groen, R., Orie, V. et al. Analysis of macaque BTN3A genes and transcripts in the extended MHC: conserved orthologs of human γδ T cell modulators. Immunogenetics 71, 545–559 (2019). https://doi.org/10.1007/s00251-019-01126-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-019-01126-9