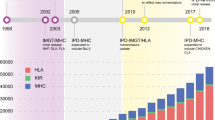
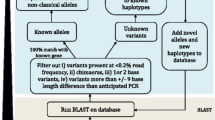
Abstract
Significant progress has been made over the last decade in defining major histocompatibility complex (MHC) diversity at the nucleotide, allele, haplotype, diplotype, and population levels in many non-human species. Much of this progress has been driven by the increased availability and reduced costs associated with nucleotide sequencing technologies. This report provides an update on the activities of the comparative MHC nomenclature committee which is a standing committee of both the International Society for Animal Genetics (ISAG) and the International Union of Immunological Societies (IUIS) where it operates under the umbrella of the Veterinary Immunology Committee (VIC). A previous report from this committee in 2006 defined the role of the committee in providing guidance in the development of a standardized nomenclature for genes and alleles at MHC loci in non-human species. It described the establishment of the Immuno Polymorphism Database, IPD-MHC, which continues to provide public access to high quality MHC sequence data across a range of species. In this report, guidelines for the continued development of a universal MHC nomenclature framework are described, summarizing the continued development of each species section within the IPD-MHC project.
Similar content being viewed by others
References
Ballingall KT, Steele P, Lantier I, Cotelli M, Todd H, Lopez G, Martin E, Lantier F (2015) An ancient inter-locus recombination increases class II MHC DQA diversity in sheep and other bovidae. Anim Genet 46:333–336
Ballingall KT, Lantier I, Todd H, Lantier F, Rocchi M (2018) Structural and functional diversity arising from intra and inter haplotype combinations of duplicated DQA and B loci within the Ovine MHC. Immunogenetics 70:257–269
Bingulac-Popovic J, Figueroa F, Sato A, Talbot WS, Johnson SL, Gates M, Postlethwait JH, Klein J (1997) Mapping of mhc class I and class II regions to different linkage groups in the zebrafish, Danio rerio. Immunogenetics 46:129–134
de Groot NG, Otting N, Robinson J, Blancher A, Lafont BAP, Marsh SGE, O'Connor DH, Shiina T, Walter L, Watkins DI, Bontrop RE (2012) Nomenclature report on the major histocompatibility complex genes and alleles of Great Ape, Old and New World monkey species. Immunogenetics 64:615–631
Ellis SA, Bontrop RE, Antczak DF, Ballingall K, Davies CJ, Kaufman J, Kennedy LJ, Robinson J, Smith DM, Stear MJ, Stet RJM, Waller MJ, Walter L, Marsh SGE (2006) ISAG/IUIS-VIC comparative MHC nomenclature committee report, 2005. Immunogenetics 57:953–958
Grimholt U (2016) MHC and evolution in teleosts. Biology 5:1. https://doi.org/10.3390/biology5010006
Grimholt U, Getahun A, Hermsen T, Stet RJ (2000) The major histocompatibility class II alpha chain in salmonid fishes. Dev Comp Immunol 24:751–763
Grimholt U, Tsukamoto K, Azuma T, Leong J, Koop BF, Dijkstra JM (2015) A comprehensive analysis of teleost MHC class I sequences. BMC Evol Biol 15:32
Hammond JA, Marsh SG, Robinson J, Davies CJ, Stear MJ, Ellis SA (2012) Cattle MHC nomenclature: is it possible to assign sequences to discrete class I genes? Immunogenetics 64:475–480
Ho CS, Lunney JK, Ando A, Rogel-Gaillard C, Lee JH, Schook LB, Smith DM (2009) Nomenclature for factors of the SLA system, update 2008. Tissue Antigens 73:307–315
Hosomichi K, Miller MM, Goto RM, Wang Y, Suzuki S, Kulski JK, Nishibori M, Inoko H, Hanzawa K, Shiina T (2008) Contribution of mutation, recombination, and gene conversion to chicken MHC-B haplotype diversity. J Immunol 181:3393–3399
Hurt P, Walter L, Sudbrak R, Klages S, Müller I, Shiina T, Inoko H, Lehrach H, Günther E, Reinhardt R, Himmelbauer H (2004) The genomic sequence and comparative analysis of the rat major histocompatibility complex. Genome Res 14:631–639
Klein J (1986) Natural history of the major histocompatibility complex. Published by J Wiley and Sons, New York
Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER, Jones PP, Parham P, Wakeland EK, Watkins DI (1990) Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics 31:217–219
Lima-Rosa CA, Canal CW, Streck AF, Freitas LB, Delgado-Cañedo A, Bonatto SL, Salzano FM (2004) B-F DNA sequence variability in Brazilian (blue-egg Caipira) chickens. Anim Genet 35:278–284
Livant EJ, Brigati JR, Ewald SJ (2004) Diversity and locus specificity of chicken MHC B class I sequences. Anim Genet 35:18–27
Maccari G, Robinson J, Ballingall K, Guethlein L, Grimholt U, Kaufmann J, Ho C-K, de Groot N, Flicek P, Bontrop R, Hammond J, Marsh S (2017) IPD-MHC 2.0: an improved inter-species database for the study of the major histocompatibility complex. Nucleic Acids Res 45(Database issue):D860–D864. https://doi.org/10.1093/nar/gkw1050
Maccari G, Robinson J, Bontrop RE, Otting N, de Groot NG, Ho C-S, Ballingall KT, Marsh SGE, Hammond JA (2018) IPD-MHC: nomenclature requirements for the non-human major histocompatibility complex in the next-generation sequencing era. Immunogenetics. https://doi.org/10.1007/s00251-018-1072-4
Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Fernández-Viña M, Geraghty DE, Holdsworth R, Hurley CK, Lau M, Lee KW, Mach B, Maiers M, Mayr WR, Müller CR, Parham P, Petersdorf EW, Sasazuki T, Strominger JL, Svejgaard A, Terasaki PI, Tiercy JM, Trowsdale J (2010) Nomenclature for factors of the HLA system, 2010. Tissue Antigens 75:291–455
Miller MM, Bacon LD, Hala K, Hunt HD, Ewald SJ, Kaufman J, Zoorob R, Briles WE (2004) Nomenclature for the chicken major histocompatibility (B and Y) complex. Immunogenetics 56:261–279
Miller D, Tallmadge RL, Binns M, Zhu B, Mohamoud YA, Ahmed A, Brooks SA, Antczak DF (2016) Polymorphism at expressed DQ and DR loci in five common equine MHC haplotypes. Immunogenetics 69:145–156
Miltiadou D, Ballingall KT, Ellis SA, Russell GC, McKeever DJ (2005) Haplotype characterization of transcribed ovine major histocompatibility complex (MHC) class I genes. Immunogenetics 57:499–509
Nonaka MI, Aizawa K, Mitani H, Bannai HP, Nonaka M (2011) Retained orthologous relationships of the MHC Class I genes during euteleost evolution. Mol Biol Evol 28:3099–3112
Robinson J, Waller MJ, Stoehr P, Marsh SGE (2005) IPD - the Immuno Polymorphism Database. Nucleic Acids Res 33:D523–D526
Robinson J, Halliwell JA, Hayhurst JH, Flicek P, Parham P, Marsh SGE (2015) The IPD and IPD-IMGT/HLA database: allele variant databases. Nucleic Acids Res 43:D423–D431
Shiina T, Dijkstra JM, Shimizu S, Watanabe A, Yanagiya K, Kiryu I, Fujiwara A, Nishida-Umehara C, Kaba Y, Hirono I et al (2005) Interchromosomal duplication of major histocompatibility complex class I regions in rainbow trout (Oncorhynchus mykiss), a species with a presumably recent tetraploid ancestry. Immunogenetics 56:878–893
Simonsen M, Hála K, Nicolaisen EM (1980) Linkage disequilibrium of MHC genes in the chicken. I. The B-F and B-G loci. Immunogenetics 10:103–112
Simonsen M, Crone M, Koch C, Hála K (1982) The MHC haplotypes of the chicken. Immunogenetics 16:513–532
Smith DM, Lunney JK, Martens GW, Ando A, Lee JH, Ho CS, Schook L, Renard C, Chardon P (2005a) Nomenclature for factors of the SLA class-I system, 2004. Tissue Antigens 65:136–149
Smith DM, Lunney JK, Ho CS, Martens GW, Ando A, Lee JH, Schook L, Renard C, Chardon P (2005b) Nomenclature for factors of the swine leukocyte antigen class II system, 2005. Tissue Antigens 66:623–639
Tallmadge RL, Lear TL, Antczak DF (2005) Genomic characterization of MHC class I genes of the horse. Immunogenetics 57:763–774
Tallmadge RL, Campbell JA, Miller DC, Antczak DF (2010) Analysis of MHC class I genes across horse MHC haplotypes. Immunogenetics 62:159–172
Wade CM, Giulotto E, Sigurdsson S, Zoli M et al (2009) Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326:865–867
WHO Nomenclature Committee (1968) Nomenclature for factors of the HL-a system. Bull World Health Organ 39:483–486
Worley K, Gillingham M, Jensen P, Kennedy LJ, Pizzari T, Kaufman J, Richardson DS (2008) Single locus typing of MHC class I and class II B loci in a population of red jungle fowl. Immunogenetics 60:233–247
Acknowledgements
The support of all those who provide their time to assign official names to MHC alleles across many different species is acknowledged. The Comparative MHC Nomenclature Committee would also like to acknowledge the Biotechnology and Biological Sciences Research Council (BBSRC) for supporting the maintenance and development of the IPD-MHC through grant BB/M011488/1 and IUIS-VIC for supporting the comparative MHC workshops at ISAG and IVIS meetings.
Funding
KB receives funding from the Scottish Government’s strategic research program and the European Union’s Horizon 2020 research and innovation program under grant agreement No. 731014. JAH and GM were also supported by the BBSRC through projects BBS/E/I/00001710, BBS/E/I/00007030, BBS/E/I/00007038, and BBS/E/I/00007039.
Author information
Authors and Affiliations
Corresponding author
Additional information
Shirley A. Ellis is no longer at The Pirbright Institute
Rights and permissions
About this article
Cite this article
Ballingall, K.T., Bontrop, R.E., Ellis, S.A. et al. Comparative MHC nomenclature: report from the ISAG/IUIS-VIC committee 2018. Immunogenetics 70, 625–632 (2018). https://doi.org/10.1007/s00251-018-1073-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-018-1073-3