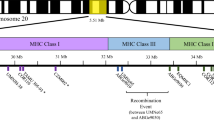
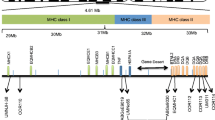
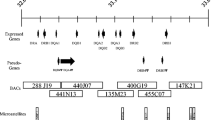
Abstract
The genomic sequences of 15 horse major histocompatibility complex (MHC) class I genes and a collection of MHC class I homozygous horses of five different haplotypes were used to investigate the genomic structure and polymorphism of the equine MHC. A combination of conserved and locus-specific primers was used to amplify horse MHC class I genes with classical and nonclassical characteristics. Multiple clones from each haplotype identified three to five classical sequences per homozygous animal and two to three nonclassical sequences. Phylogenetic analysis was applied to these sequences, and groups were identified which appear to be allelic series, but some sequences were left ungrouped. Sequences determined from MHC class I heterozygous horses and previously described MHC class I sequences were then added, representing a total of ten horse MHC haplotypes. These results were consistent with those obtained from the MHC homozygous horses alone, and 30 classical sequences were assigned to four previously confirmed loci and three new provisional loci. The nonclassical genes had few alleles and the classical genes had higher levels of allelic polymorphism. Alleles for two classical loci with the expected pattern of polymorphism were found in the majority of haplotypes tested, but alleles at two other commonly detected loci had more variation outside of the hypervariable region than within. Our data indicate that the equine major histocompatibility complex is characterized by variation in the complement of class I genes expressed in different haplotypes in addition to the expected allelic polymorphism within loci.






Similar content being viewed by others
Abbreviations
- ARS:
-
Antigen recognition site
- BAC:
-
Bacterial artificial chromosome
- CTL:
-
Cytotoxic T lymphocyte
- ELA:
-
Equine leukocyte antigen
- HLA:
-
Human leukocyte antigen
- MHC:
-
Major histocompatibility complex
- NK:
-
Natural killer
- UTR:
-
Untranslated region
References
Ando A, Kawata H, Shigenari A, Anzai T, Ota M, Katsuyama Y, Sada M, Goto R, Takeshima SN, Aida Y, Iwanaga T, Fujimura N, Suzuki Y, Gojobori T, Inoko H (2003) Genetic polymorphism of the swine major histocompatibility complex (SLA) class I genes, SLA-1, -2 and -3. Immunogenetics 55:583–593
Antczak DF, Bright SM, Remick LH, Bauman BE (1982) Lymphocyte alloantigens of the horse. I. Serologic and genetic studies. Tissue Antigens 20:172–187
Antczak DF, Bailey E, Barger B, Guerin G, Lazary S, McClure J, Mottironi VD, Symons R, Templeton J, Varewyck H (1986) Joint report of the Third International Workshop on Lymphocyte Alloantigens of the Horse, Kennett Square, Pennsylvania, 25–27 April 1984. Anim Genet 17:363–373
Babiuk S, Horseman B, Zhang C, Bickis M, Kusalik A, Schook LB, Abrahamsen MS, Pontarollo R (2007) BoLA class I allele diversity and polymorphism in a herd of cattle. Immunogenetics 59:167–176
Barbis DP, Maher JK, Stanek J, Klaunberg BA, Antczak DF (1994) Horse cDNA clones encoding two MHC class I genes. Immunogenetics 40:163
Birch J, Codner G, Guzman E, Ellis SA (2008) Genomic location and characterisation of nonclassical MHC class I genes in cattle. Immunogenetics 60:267–273
Bjorkman PJ, Parham P (1990) Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem 59:253–288
Carpenter S, Baker JM, Bacon SJ, Hopman T, Maher J, Ellis SA, Antczak DF (2001) Molecular and functional characterization of genes encoding horse MHC class I antigens. Immunogenetics 53:802–809
Cereb N, Yang SY (1994) The regulatory complex of HLA class I promoters exhibits locus-specific conservation with limited allelic variation. J Immunol 152:3873–3883
Cereb N, Maye P, Lee S, Kong Y, Yang SY (1995) Locus-specific amplification of HLA class I genes from genomic DNA: locus-specific sequences in the first and third introns of HLA-A, -B, and -C alleles. Tissue Antigens 45:1–11
Chardon P, Renard C, Vaiman M (1999) The major histocompatibility complex in swine. Immunol Rev 167:179–192
Chung C, Leib SR, Fraser DG, Ellis SA, McGuire TC (2003) Novel classical MHC class I alleles identified in horses by sequencing clones of reverse transcription-PCR products. Eur J Immunogenet 30:387–396
Ellis S (2004) The cattle major histocompatibility complex: is it unique? Vet Immunol Immunopathol 102:1–8
Ellis SA, Martin AJ, Holmes EC, Morrison WI (1995) At least four MHC class I genes are transcribed in the horse: phylogenetic analysis suggests an unusual evolutionary history for the MHC in this species. Eur J Immunogenet 22:249–260
Ellis SA, Holmes EC, Staines KA, Smith KB, Stear MJ, McKeever DJ, MacHugh ND, Morrison WI (1999) Variation in the number of expressed MHC genes in different cattle class I haplotypes. Immunogenetics 50:319–328
Ellis SA, Morrison WI, MacHugh ND, Birch J, Burrells A, Stear MJ (2005) Serological and molecular diversity in the cattle MHC class I region. Immunogenetics 57:601–606
Ellis SA, Bontrop RE, Antczak DF, Ballingall K, Davies CJ, Kaufman J, Kennedy LJ, Robinson J, Smith DM, Stear MJ, Stet RJ, Waller MJ, Walter L, Marsh SG (2006) ISAG/IUIS-VIC comparative MHC Nomenclature committee report, 2005. Immunogenetics 57:953–958
Germain RN, Margulies DH (1993) The biochemistry and cell biology of antigen processing and presentation. Annu Rev Immunol 11:403–450
Gu X, Nei M (1999) Locus specificity of polymorphic alleles and evolution by a birth-and-death process in mammalian MHC genes. Mol Biol Evol 16:147–156
Gustafson AL, Tallmadge RL, Ramlachan N, Miller D, Bird H, Antczak DF, Raudsepp T, Chowdhary BP, Skow LC (2003) An ordered BAC contig map of the equine major histocompatibility complex. Cytogenet Genome Res 102:189–195
Holmes EC, Ellis SA (1999) Evolutionary history of MHC class I genes in the mammalian order Perissodactyla. J Mol Evol 49:316–324
Hughes AL, Nei M (1988) Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335:167–170
Johnson DR (2000) Differential expression of human major histocompatibility class I loci: HLA-A, -B, and -C. Hum Immunol 61:389–396
Joly E, Leong L, Coadwell W, Clarkson C, Butcher G (1996) The rat MHC haplotype RT1c expresses two classical class I molecules. J Immunol 157:1551–1558
Kaufman J, Jacob J, Shaw I, Walker B, Milne S, Beck S, Salomonsen J (1999) Gene organisation determines evolution of function in the chicken MHC. Immunol Rev 167:101–117
Kirisits MJ, Kunz HW, Hassett AL, Gill TJ3rd (1992) Genomic DNA sequence and organization of a TL-like gene in the grc-G/C region of the rat. Immunogenetics 35:365–377
Koller BH, Sidwell B, DeMars R, Orr HT (1984) Isolation of HLA locus-specific DNA probes from the 3′-untranslated region. Proc Natl Acad Sci U S A 81:5175–5178
Korber B (2000) HIV signature and sequence variation analysis. In: Rodrigo AG, Learn GH (eds) Computational analysis of HIV molecular sequences. The Netherlands, Kluwer Academic, pp 55–72
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Kydd JH, Davis-Poynter NJ, Birch J, Hannant D, Minke J, Audonnet JC, Antczak DF, Ellis SA (2006) A molecular approach to the identification of cytotoxic T-lymphocyte epitopes within equine herpesvirus 1. J Gen Virol 87:2507–2515
Lanier LL, Corliss B, Phillips JH (1997) Arousal and inhibition of human NK cells. Immunol Rev 155:145–154
Lau P, Lorenzi R, Joly E (2000) Comparison of RT-BM1 sequences from six different rat major histocompatibility complex haplotypes reveals limited variation, and alternate splicing in the 3′ untranslated region. Immunogenetics 51:148–153
Lawlor DA, Zemmour J, Ennis PD, Parham P (1990) Evolution of class-I MHC genes and proteins: from natural selection to thymic selection. Annu Rev Immunol 8:23–63
Lazary S, Antczak DF, Bailey E, Bell TK, Bernoco D, Byrns G, McClure JJ (1988) Joint report of the Fifth International Workshop on Lymphocyte Alloantigens of the Horse, Baton Rouge, Louisiana, 31 October–1 November 1987. Anim Genet 19:447–456
Le Bouteiller P (1994) HLA class I chromosomal region, genes, and products: facts and questions. Crit Rev Immunol 14:89–129
McGuire TC, Leib SR, Mealey RH, Fraser DG, Prieur DJ (2003) Presentation and binding affinity of equine infectious anemia virus CTL envelope and matrix protein epitopes by an expressed equine classical MHC class I molecule. J Immunol 171:1984–1993
Mealey RH, Lee JH, Leib SR, Littke MH, McGuire TC (2006) A single amino acid difference within the alpha-2 domain of two naturally occurring equine MHC class I molecules alters the recognition of Gag and Rev epitopes by equine infectious anemia virus-specific CTL. J Immunol 177:7377–7390
Miltiadou D, Ballingall KT, Ellis SA, Russell GC, McKeever DJ (2005) Haplotype characterization of transcribed ovine major histocompatibility complex (MHC) class I genes. Immunogenetics 57:499–509
Moore KW, Sher BT, Sun YH, Eakle KA, Hood L (1982) DNA sequence of a gene encoding a BALB/c mouse Ld transplantation antigen. Science 215:679–682
Mungall AJ, Palmer SA, Sims SK, Edwards CA, Ashurst JL, Wilming L, Jones MC, Horton R, Hunt SE, Scott CE, Gilbert JG, Clamp ME, Bethel G, Milne S, Ainscough R, Almeida JP, Ambrose KD, Andrews TD, Ashwell RI, Babbage AK et al (2003) The DNA sequence and analysis of human chromosome 6. Nature 425:805–811
Ober C, Aldrich C, Rosinsky B, Robertson A, Walker MA, Willadsen S, Verp MS, Geraghty DE, Hunt JS (1998) HLA-G1 protein expression is not essential for fetal survival. Placenta 19:127–132
Otting N, Heijmans CM, Noort RC, de Groot NG, Doxiadis GG, van Rood JJ, Watkins DI, Bontrop RE (2005) Unparalleled complexity of the MHC class I region in rhesus macaques. Proc Natl Acad Sci U S A 102:1626–1631
Parham P, Lomen CE, Lawlor DA, Ways JP, Holmes N, Coppin HL, Salter RD, Wan AM, Ennis PD (1988) Nature of polymorphism in HLA-A, -B, and -C molecules. Proc Natl Acad Sci USA 85:4005–4009
Parham P, Lawlor DA, Lomen CE, Ennis PD (1989) Diversity and diversification of HLA-A, B, C alleles. J Immunol 142:3937–3950
Parham P, Adams EJ, Arnett KL (1995) The origins of HLA-A, B, C polymorphism. Immunol Rev 143:141–180
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Smith DM, Lunney JK, Martens GW, Ando A, Lee JH, Ho CS, Schook L, Renard C, Chardon P (2005) Nomenclature for factors of the SLA class-I system, 2004. Tissue Antigens 65:136–149
Steinmetz M, Frelinger JG, Fisher D, Hunkapiller T, Pereira D, Weissman SM, Uehara H, Nathenson S, Hood L (1981) Three cDNA clones encoding mouse transplantation antigens: homology to immunoglobulin genes. Cell 24:125–134
Stroynowski I (1990) Molecules related to class-I major histocompatibility complex antigens. Annu Rev Immunol 8:501–530
Swelsen WT, Voorter CE, van den Berg-Loonen EM (2003) Polymorphism of intron 4 in HLA-A, -B and -C genes. Tissue Antigens 61:475–483
Tallmadge RL, Lear TL, Antczak DF (2005) Genomic characterization of MHC class I genes of the horse. Immunogenetics 57:763–774
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tanaka-Matsuda M, Ando A, Rogel-Gaillard C, Chardon P, Uenishi H (2009) Difference in number of loci of swine leukocyte antigen classical class I genes among haplotypes. Genomics 93:261–273
Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, Wilson MJ (2001) The genomic context of natural killer receptor extended gene families. Immunol Rev 181:20–38
Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, Blocker H, Distl O, Edgar RC, Garber M, Leeb T, Mauceli E, MacLeod JN, Penedo MC, Raison JM, Sharpe T, Vogel J, Andersson L, Antczak DF, Biagi T, Binns MM, Chowdhary BP, Coleman SJ, Della Valle G, Fryc S, Guerin G, Hasegawa T, Hill EW, Jurka J, Kiialainen A, Lindgren G, Liu J, Magnani E, Mickelson JR, Murray J, Nergadze SG, Onofrio R, Pedroni S, Piras MF, Raudsepp T, Rocchi M, Roed KH, Ryder OA, Searle S, Skow L, Swinburne JE, Syvanen AC, Tozaki T, Valberg SJ, Vaudin M, White JR, Zody MC, Lander ES, Lindblad-Toh K (2009) Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326:865–867
Wu TT, Kabat EA (1970) An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J Exp Med 132:211–250
Acknowledgments
This research was funded by NIH grants HD-34086, HD-15799, and HD-049545, the Harry M. Zweig Memorial Fund for Equine Research, the Dorothy Russell Havemeyer Foundation, Inc., and the USDA Regional Grants Program. We thank Dr. Shirley Ellis, Dr. Ashley Moffett, Dr. John Trowsdale, and Dr. Stephan Beck for helpful discussions and Dr. Robert H. Mealey of Washington State University for generously providing samples from horses of known ELA serotypes.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Phylogenetic tree of horse nonclassical MHC class I cDNA sequences. The tree was constructed based on the coding sequences of putative nonclassical horse MHC class I genes (Tables 1; Supplementary Table 1) excluding residues of the antigen recognition site (ARS). Locus groupings are shown on the right. Sequence names exclude the prefix “Eqca”. Groups A, C, and E refer to phylogenetic analysis by Holmes and Ellis (1999). Sequences were analyzed using the neighbor-joining method (Saitou and Nei 1987) on the basis of Tamura–Nei distances (Tamura and Nei 1993) using the MEGA 3.1 program. Percent of supporting bootstrap replications (n = 1,000) are shown next to the branches. HLA-A*0101 sequence included as an outgroup to root the phylogenetic tree (PDF 12 kb)
Supplementary Figure 2
Phylogenetic tree of classical MHC class I sequences from MHC homozygous horses. Phylogenetic tree was constructed based on the A) coding sequence excluding residues of the antigen recognition site or B) 3′UTR sequence of classical horse MHC class I genes (Table 1). All of these sequences clustered with group B sequences defined by Holmes and Ellis (1999, data not shown). Allele names are followed by associated serological ELA haplotype and exclude the prefix “Eqca”. Putative ELA locus designations are shown on the right. Sequences were analyzed using the neighbor-joining method (Saitou and Nei 1987) on the basis of Tamura–Nei distances (Tamura and Nei 1993) using the MEGA 3.1 program. Percent of supporting bootstrap replications (n = 1,000) are shown next to the branches. HLA-A*0101 sequence included as an outgroup to root the phylogenetic tree. Provisional locus assignments are given in italics and followed by an asterisk (PDF 17.2 kb)
Supplementary Figure 3
Phylogenetic tree of horse MHC class I 3′UTR sequences from ten equine MHC haplotypes. The tree was constructed based on horse MHC class I 3′UTR sequences (Tables 1; Supplementary Table 1). All of these sequences clustered with group B sequences defined by Holmes and Ellis (1999). Allele names are followed by associated serological ELA haplotype and exclude the prefix “Eqca”. Putative locus assignments were shown on the right; provisional locus assignments are given in italics and followed by an asterisk. Sequences were analyzed using the neighbor-joining method (Saitou and Nei 1987) on the basis of Tamura–Nei distances (Tamura and Nei 1993) using the MEGA 3.1 program. Percent of supporting bootstrap replications (n = 1,000) are shown next to the branches. HLA-A*0101 sequence included as an outgroup to root the phylogenetic tree (PDF 15 kb)
Supplementary Figure 4
Nucleotide alignment of horse MHC class I 3′UTR sequences. Allele names are followed by associated serological ELA haplotype and exclude the prefix “Eqca”. The consensus sequence is shown at the top, dots represent identities, and dashes represent gaps inserted to optimize the alignment. Horizontal gray bars separate loci, listed on the right. Sequences are from Table 1; the reverse primer sequence is not included (PDF 18 kb)
Supplementary Figure 5
Phylogenetic tree of horse MHC class I intron 2 sequences from nonclassical loci 5, 6, and 7. The tree was constructed based on the intron 2 sequences from nonclassical loci 5, 6, and 7 (DQ145590-2, DQ083411-3), which correspond to groups A, C, and E, respectively, defined by Holmes and Ellis (1999). Allele names are followed by associated serological ELA haplotype and exclude the prefix “Eqca”. Locus assignments are shown on the right. Sequences were analyzed using the neighbor-joining method (Saitou and Nei 1987) on the basis of Tamura–Nei distances (Tamura and Nei 1993) using the MEGA 3.1 program. Percent of supporting bootstrap replications (n = 1,000) are shown next to the branches. HLA-A*0101 intron 2 sequence was included as an outgroup to root the phylogenetic tree (PDF 10 kb)
Supplementary Table 1
Previously published equine MHC class I sequences associated with serologically defined haplotypes included in this study (PDF 16 kb)
Supplementary Table 2
Oligonucleotides used for RT-PCR amplification of horse MHC class I genesa (PDF 13 kb)
Supplementary Table 3
Nomenclature for equine MHC class I alleles (PDF 32 kb)
Rights and permissions
About this article
Cite this article
Tallmadge, R.L., Campbell, J.A., Miller, D.C. et al. Analysis of MHC class I genes across horse MHC haplotypes. Immunogenetics 62, 159–172 (2010). https://doi.org/10.1007/s00251-009-0420-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-009-0420-9