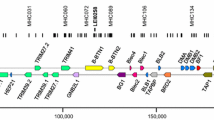
Abstract
The major histocompatibility complex (MHC) is a key genomic model region for understanding the evolution of gene families and the co-evolution between host and pathogen. To date, MHC studies have mostly focused on species from major vertebrate lineages. The evolution of MHC classical (Ia) and non-classical (Ib) genes in pigs has attracted interest because of their antigen presentation roles as part of the adaptive immune system. The pig family Suidae comprises over 18 extant species (mostly wild), but only the domestic pig has been extensively sequenced and annotated. To address this, we used a DNA-capture approach, with probes designed from the domestic pig genome, to generate MHC data for 11 wild species of pigs and their closest living family, Tayassuidae. The approach showed good efficiency for wild pigs (~80% reads mapped, ~87× coverage), compared to tayassuids (~12% reads mapped, ~4× coverage). We retrieved 145 MHC loci across both families. Phylogenetic analyses show that the class Ia and Ib genes underwent multiple duplications and diversifications before suids and tayassuids diverged from their common ancestor. The histocompatibility genes mostly form orthologous groups and there is genetic differentiation for most of these genes between Eurasian and sub-Saharan African wild pigs. Tests of selection showed that the peptide-binding region of class Ib genes was under positive selection. These findings contribute to better understanding of the evolutionary history of the MHC, specifically, the class I genes, and provide useful data for investigating the immune response of wild populations against pathogens.



Similar content being viewed by others
References
Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, Fernandez-Banet J, Billis K, Garcia-Giron C, Hourlier T, Howe KL, Kahari AK, Kokocinski F, Martin FJ, Murphy DN, Nag R, Ruffier M, Schuster M, Tang YA, Vogel J-H, White S, Zadissa A, Flicek P, Searle SMJ, Fernandez Banet J, Billis K, García Girón C, Hourlier T, Howe KL, Kähäri A, Kokocinski F, Martin FJ, Murphy DN, Nag R, Ruffier M, Schuster M, Tang YA, Vogel J-H, White S, Zadissa A, Flicek P, Searle SMJ (2016) The Ensembl Gene Annotation System. Database (Oxford) 2016:baw093. doi: https://doi.org/10.1093/database/baw093
Al Dahouk S, Nöckler K, Tomaso H, Splettstoesser WD, Jungersen G, Riber U, Petry T, Hoffmann D, Scholz HC, Hensel A, Neubauer H (2005) Seroprevalence of brucellosis, tularemia, and yersiniosis in wild boars (Sus scrofa) from north-eastern Germany. J Vet Med B Infect Dis Vet Public Health 52(10):444–455. https://doi.org/10.1111/j.1439-0450.2005.00898.x
Alcaide M, Muñoz J, Martínez-de la Puente J, Soriguer R, Figuerola J (2014) Extraordinary MHC class II B diversity in a non-passerine, wild bird: the Eurasian coot Fulica atra (Aves: Rallidae). Ecol Evol 4(6):688–698. https://doi.org/10.1002/ece3.974
Allen RL, Hogan L (2013) Non-classical MHC class I molecules (MHC-Ib). eLS. John Wiley & Sons, Ltd, Chichester, pp 1–12
Amadou C (1999) Evolution of the MHC class I region: the framework hypothesis. Immunogenetics 49(4):362–367. https://doi.org/10.1007/s002510050507
Ando A, Chardon P (2006) Gene organization and polymorphism of the swine major histocompatibility complex. Anim Sci J 77(2):127–137. https://doi.org/10.1111/j.1740-0929.2006.00331.x
Anzai T, Shiina T, Kimura N, Yanagiya K, Kohara S, Shigenari A, Yamagata T, Kulski JK, Naruse TK, Fujimori Y, Fukuzumi Y, Yamazaki M, Tashiro H, Iwamoto C, Umehara Y, Imanishi T, Meyer A, Ikeo K, Gojobori T, Bahram S, Inoko H (2003) Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence. Proc Natl Acad Sci U S A 100(13):7708–7713. https://doi.org/10.1073/pnas.1230533100
Ballingall KT, McKeever DJ (2005) Conservation of promoter, coding and intronic regions of the nonclassical MHC class II DYA gene suggests evolution under functional constraints. Anim Genet 36(3):237–239. https://doi.org/10.1111/j.1365-2052.2005.01281.x
Barbisan F, Savio C, Bertorelle G, Patarnello T, Congiu L (2009) Duplication polymorphism at MHC class II DRB1 locus in the wild boar (Sus scrofa). Immunogenetics 61(2):145–151. https://doi.org/10.1007/s00251-008-0339-6
Barrett LW, Fletcher S, Wilton SD (2013) Untranslated gene regions and other non-coding elements regulation of Eukaryotic gene expression. Series: Springer Briefs in Biochemistry and Molecular Biology, Springer, Basel, pp 1–56
Borghans JAM, Beltman JB, de Boer RJ (2004) MHC polymorphism under host-pathogen coevolution. Immunogenetics 55(11):732–739. https://doi.org/10.1007/s00251-003-0630-5
Brinton RD, Thompson RF, Foy MR, Baudry M, Wang J, Finch CE, Morgan TE, Pike CJ, Mack WJ, Stanczyk FZ, Nilsen J (2008) Progesterone receptors: form and function in brain. Front Neuroendocrinol 29(2):313–339. https://doi.org/10.1016/j.yfrne.2008.02.001
Buckley BA (2007) Comparative environmental genomics in non-model species: using heterologous hybridization to DNA-based microarrays. J Exp Biol 210(9):1602–1606. https://doi.org/10.1242/jeb.002402
Burbano HA, Hodges E, Green RE, Briggs AW, Krause J, Meyer M, Good JM, Maricic T, Johnson PLF, Xuan Z, Rooks M, Bhattacharjee A, Brizuela L, Albert FW, de la Rasilla M, Fortea J, Rosas A, Lachmann M, Hannon GJ, Pääbo S (2010) Targeted investigation of the Neandertal genome by array-based sequence capture. Science 328(5979):723–725. https://doi.org/10.1126/science.1188046
Burri R, Hirzel HN, Salamin N, Roulin A, Fumagalli L (2008) Evolutionary patterns of MHC class II B in owls and their implications for the understanding of avian MHC evolution. Mol Biol Evol 25(6):1180–1191. https://doi.org/10.1093/molbev/msn065
Carver EA, Stubbs L (1997) Zooming in on the human–mouse comparative map: genome conservation re-examined on a high-resolution scale. Genome Res 7(12):1123–1137. https://doi.org/10.1101/gr.7.12.1123
Castley ASL, Martinez OP (2012) Molecular analysis of complement component C4 gene copy number. In: Christiansen FT, Tait BD (eds) Immunogenetics. Humana Press, Totowa, pp 159–171. https://doi.org/10.1007/978-1-61779-842-9_9
Chiovaro F, Chiquet-Ehrismann R, Chiquet M (2015) Transcriptional regulation of tenascin genes. Cell Adh Migr 9:34–47. https://doi.org/10.1080/19336918.2015.1008333
Choo KH, Vissel B, Nagy A, Earle E, Kalitsis P (1991) A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res 19(6):1179–1182. https://doi.org/10.1093/nar/19.6.1179
Costard S, Wieland B, De GW, Jori F, Rowlands R, Vosloo W, Roger F, Pfeiffer DU, Dixon LK, Royal T, College V, Lane H, Al H, de Glanville W, Jori F, Rowlands R, Vosloo W, Roger F, Pfeiffer DU, Dixon LK (2009) African swine fever: how can global spread be prevented? Philos Trans R Soc Lond Ser B Biol Sci 364(1530):2683–2696. https://doi.org/10.1098/rstb.2009.0098
Crew MD, Phanavanh B, Garcia-Borges CN (2004) Sequence and mRNA expression of nonclassical SLA class I genes SLA-7 and SLA-8. Immunogenetics 56(2):111–114. https://doi.org/10.1007/s00251-004-0676-z
Cummings N, King R, Rickers A, Kaspi A, Lunke S, Haviv I, Jowett JBM (2010) Combining target enrichment with barcode multiplexing for high throughput SNP discovery. BMC Genomics 11(1):641. https://doi.org/10.1186/1471-2164-11-641
Day WH, McMorris FR (1992) Critical comparison of consensus methods for molecular sequences. Nucleic Acids Res 20(5):1093–1099. https://doi.org/10.1093/nar/20.5.1093
Deakin JE, Papenfuss AT, Belov K, Cross JGR, Coggill P, Palmer S, Sims S, Speed TP, Beck S, Graves JAM (2006) Evolution and comparative analysis of the MHC class III inflammatory region. BMC Genomics 7(1):281. https://doi.org/10.1186/1471-2164-7-281
Dixon LK, Abrams CC, Bowick G, Goatley LC, Kay-Jackson PC, Chapman D, Liverani E, Nix R, Silk R, Zhang F (2004) African swine fever virus proteins involved in evading host defence systems. Vet Immunol Immunopathol 100(3-4):117–134. https://doi.org/10.1016/j.vetimm.2004.04.002
Drake JA, Bird C, Nemesh J, Thomas DJ, Newton-Cheh C, Reymond A, Excoffier L, Attar H, Antonarakis SE, Dermitzakis ET, Hirschhorn JN (2006) Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat Genet 38(2):223–227. https://doi.org/10.1038/ng1710
Ebbert MTW, Wadsworth ME, Staley LA, Hoyt KL, Pickett B, Miller J, Duce J, Neuroimaging D, Kauwe JSK, Ridge PG (2016) Evaluating the necessity of PCR duplicate removal from next-generation sequencing data and a comparison of approaches. BMC Bioinformatics 17(S7):239. https://doi.org/10.1186/s12859-016-1097-3
Emes RD, Goodstadt L, Winter EE, Ponting CP (2003) Comparison of the genomes of human and mouse lays the foundation of genome zoology. Hum Mol Genet 12(7):701–709. https://doi.org/10.1093/hmg/ddg078
Essler SE, Ertl W, Deutsch J, Ruetgen BC, Groiss S, Stadler M, Wysoudil B, Gerner W, Ho C-S, Saalmueller A (2013) Molecular characterization of swine leukocyte antigen gene diversity in purebred Pietrain pigs. Anim Genet 44(2):202–205. https://doi.org/10.1111/j.1365-2052.2012.02375.x
Fay JC, Wyckoff GJ, CI W (2001) Positive and negative selection on the human genome. Genetics 158(1):1227–1234. https://doi.org/10.1016/s0378-1119(99)00294-2
Fowler ME (1996) Husbandry and diseases of captive wild swine and peccaries. Rev Sci Tech 15(1):141–154. 10.20506/rst.15.1.913
Frazer KA, Elnitski L, Church DM, Dubchak I, Hardison RC (2003) Cross-species sequence comparisons: a review of methods and available resources. Genome Res 13(1):1–12. https://doi.org/10.1101/gr.222003
Gieling ET, Nordquist RE, van der Staay FJ (2011) Assessing learning and memory in pigs. Anim Cogn 14(2):151–173. https://doi.org/10.1007/s10071-010-0364-3
Gongora J, Fleming P, Spencer PBS, Mason R, Garkavenko O, Meyer JN, Droegemueller C, Lee JH, Moran C (2004) Phylogenetic relationships of Australian and New Zealand feral pigs assessed by mitochondrial control region sequence and nuclear GPIP genotype. Mol Phylogenet Evol 33(2):339–348. https://doi.org/10.1016/j.ympev.2004.06.004
Gongora J, Cuddahee RE, Nascimento FF Do, Palgrave CJ, Lowden S, Ho SYW, Simond D, Damayanti CS, White DJ, Tay WT, Randi E, Klingel H, Rodrigues-Zarate CJ, Allen K, Moran C, Larson G (2011) Rethinking the evolution of extant sub-Saharan African suids (Suidae, Artiodactyla). Zool Scr 40:327–335. https://doi.org/10.1111/j.1463-6409.2011.00480.x
Gongora J, Groves C, Meijaard E (2017) Evolutionary relationships and taxonomy of suidae and tayassuidae. In: Melleti M, Meijaard E (eds) Ecology, conservation and management of wild pigs and peccaries. Cambridge University Press, pp 1–19. https://doi.org/10.1017/9781316941232.003
Groenen MAM, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, Rogel-Gaillard C, Park C, Milan D, Megens H-J, Li S, Larkin DM, Kim H, Frantz LAF, Caccamo M, Ahn H, Aken BL, Anselmo A, Anthon C, Auvil L, Badaoui B, Beattie CW, Bendixen C, Berman D, Blecha F, Blomberg J, Bolund L, Bosse M, Botti S, Bujie Z, Bystrom M, Capitanu B, Carvalho-Silva D, Chardon P, Chen C, Cheng R, Choi S-H, Chow W, Clark RC, Clee C, Crooijmans RPMA, Dawson HD, Dehais P, de Sapio F, Dibbits B, Drou N, Z-Q D, Eversole K, Fadista J, Fairley S, Faraut T, Faulkner GJ, Fowler KE, Fredholm M, Fritz E, Gilbert JGR, Giuffra E, Gorodkin J, Griffin DK, Harrow JL, Hayward A, Howe K, Z-L H, Humphray SJ, Hunt T, Hornshøj H, Jeon J-T, Jern P, Jones M, Jurka J, Kanamori H, Kapetanovic R, Kim J, Kim J-H, Kim K-W, Kim T-H, Larson G, Lee K, Lee K-T, Leggett R, Lewin HA, Li Y, Liu W, Loveland JE, Lu Y, Lunney JK, Ma J, Madsen O, Mann K, Matthews L, McLaren S, Morozumi T, Murtaugh MP, Narayan J, Truong Nguyen D, Ni P, S-J O, Onteru S, Panitz F, Park E-W, Park H-S, Pascal G, Paudel Y, Perez-Enciso M, Ramirez-Gonzalez R, Reecy JM, Rodriguez-Zas S, Rohrer GA, Rund L, Sang Y, Schachtschneider K, Schraiber JG, Schwartz J, Scobie L, Scott C, Searle S, Servin B, Southey BR, Sperber G, Stadler P, Sweedler JV, Tafer H, Thomsen B, Wali R, Wang J, Wang J, White S, Xu X, Yerle M, Zhang G, Zhang J, Zhang J, Zhao S, Rogers J, Churcher C, Schook LB (2012) Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491(7424):393–398. https://doi.org/10.1038/nature11622
Harakalova M, Mokry M, Hrdlickova B, Renkens I, Duran K, van Roekel H, Lansu N, van Roosmalen M, de Bruijn E, Nijman IJ, Kloosterman WP, Cuppen E (2011) Multiplexed array-based and in-solution genomic enrichment for flexible and cost-effective targeted next-generation sequencing. Nat Protoc 6(12):1870–1886. https://doi.org/10.1038/nprot.2011.396
Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22(2):160–174. https://doi.org/10.1007/BF02101694
Ho CS, Lunney JK, Lee JH, Franzo-Romain MH, Martens GW, Rowland RRR, Smith DM (2010) Molecular characterization of swine leucocyte antigen class II genes in outbred pig populations. Anim Genet 41(4):428–432. https://doi.org/10.1111/j.1365-2052.2010.02019.x
Hodges E, Rooks M, Xuan Z, Bhattacharjee A, Benjamin Gordon D, Brizuela L, Richard McCombie W, Hannon GJ (2009) Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nat Protoc 4(6):960–974. https://doi.org/10.1038/nprot.2009.68
Hoppman-Chaney N, Peterson LM, Klee EW, Middha S, Courteau LK, Ferber MJ (2010) Evaluation of oligonucleotide sequence capture arrays and comparison of next-generation sequencing platforms for use in molecular diagnostics. Clin Chem 56(8):1297–1306. https://doi.org/10.1373/clinchem.2010.145441
Hu R, Lemonnier G, Bourneuf E, Vincent-Naulleau S, Rogel-Gaillard C (2011) Transcription variants of SLA-7, a swine non classical MHC class I gene. BMC Proc 5(Suppl 4):S10. https://doi.org/10.1186/1753-6561-5-S4-S10
Hughes AL, Yeager M (1998) Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet 32(1):415–435. https://doi.org/10.1146/annurev.genet.32.1.415
Illumina (2009) Sequencing analysis software—user guide, pp 1–176
Irvine AD, Irwin McLean WH (2006) Breaking the (un)sound barrier: filaggrin is a major gene for atopic dermatitis. J Invest Dermatol 126(6):1200–1202. https://doi.org/10.1038/sj.jid.5700365
Janova E, Matiasovic J, Vahala J, Vodicka R, van Dyk E, Horin P (2009) Polymorphism and selection in the major histocompatibility complex DRA and DQA genes in the family Equidae. Immunogenetics 61(7):513–527. https://doi.org/10.1007/s00251-009-0380-0
Jaratlerdsiri W, Isberg SR, Higgins DP, Gongora J (2012) MHC class I of saltwater crocodiles (Crocodylus porosus): polymorphism and balancing selection. Immunogenetics 64(11):825–838. https://doi.org/10.1007/s00251-012-0637-x
Jaratlerdsiri W, Isberg SR, Higgins DP, Ho SYW, Salomonsen J, Skjodt K, Miles LG, Gongora J (2014) Evolution of MHC class I in the order Crocodylia. Immunogenetics 66(1):53–65. https://doi.org/10.1007/s00251-013-0746-1
Johns TG, Bernard CCA (1999) The structure and function of myelin oligodendrocyte glycoprotein. J Neurochem 72(1):1–9. https://doi.org/10.1046/j.1471-4159.1999.0720001.x
Joosten SA, Sullivan LC, Ottenhoff THM (2016) Characteristics of HLA-E restricted T-cell responses and their role in infectious diseases. J Immunol Res 2016:1–11. https://doi.org/10.1155/2016/2695396
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780. https://doi.org/10.1093/molbev/mst010
Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO (2006) Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol 6(1):29. https://doi.org/10.1186/1471-2148-6-29
Kelley J, Walter L, Trowsdale J (2005) Comparative genomics of major histocompatibility complexes. Immunogenetics 56(10):683–695. https://doi.org/10.1007/s00251-004-0717-7
Kersten S, Gronemeyer H, Noy N (1997) The DNA binding pattern of the retinoid X receptor is regulated by ligand-dependent modulation of its oligomeric state. J Biol Chem 272(19):12771–12777. https://doi.org/10.1074/jbc.272.19.12771
Kirk LM, Ti SW, Bishop HI, Orozco-Llamas M, Pham M, Trimmer JS, Díaz E (2016) Distribution of the SynDIG4/proline-rich transmembrane protein 1 in rat brain. J Comp Neurol 524(11):2266–2280. https://doi.org/10.1002/cne.23945
Kloch A, Babik W, Bajer A, Siński E, Radwan J (2010) Effects of an MHC-DRB genotype and allele number on the load of gut parasites in the bank vole Myodes glareolus. Mol Ecol 19(Suppl 1):255–265. https://doi.org/10.1111/j.1365-294X.2009.04476.x
Krishnan BR, Jamry I, Chaplin DD (1995) Feature mapping of the HLA class I region: localization of the POU5F1 and TCF19 genes. Genomics 30(1):53–58. https://doi.org/10.1006/geno.1995.0008
Kulski JK (2004) Rhesus macaque class I duplicon structures, organization, and evolution within the alpha block of the major histocompatibility complex. Mol Biol Evol 21(11):2079–2091. https://doi.org/10.1093/molbev/msh216
Kulski JK, Shiina T, Anzai T, Kohara S, Inoko H (2002) Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol Rev 190(1):95–122. https://doi.org/10.1034/j.1600-065X.2002.19008.x
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Kusza S, Flori L, Gao Y, Teillaud A, Hu R, Lemonnier G, Bosze Z, Bourneuf E, Vincent-Naulleau S, Rogel-Gaillard C (2011) Transcription specificity of the class Ib genes SLA-6, SLA-7 and SLA-8 of the swine major histocompatibility complex and comparison with class Ia genes. Anim Genet 42(5):510–520. https://doi.org/10.1111/j.1365-2052.2010.02170.x
Lê S, Josse J, Husson F (2008) FactoMineR: an R package for multivariate analysis. J Stat Softw 25(1):1–18. 10.18637/jss.v025.i01
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Loughner CL, Bruford EA, McAndrews MS, Delp EE, Swamynathan SSK, Swamynathan SSK (2016) Organization, evolution and functions of the human and mouse Ly6/uPAR family genes. Hum Genomics 10(1):10. https://doi.org/10.1186/s40246-016-0074-2
Luetkemeier ES, Malhi RS, Beever JE, Schook LB (2009) Diversification of porcine MHC class II genes: evidence for selective advantage. Immunogenetics 61(2):119–129. https://doi.org/10.1007/s00251-008-0348-5
Lunney JK, Ho CS, Wysocki M, Smith DM (2009) Molecular genetics of the swine major histocompatibility complex, the SLA complex. Dev Comp Immunol 33(3):362–374. https://doi.org/10.1016/j.dci.2008.07.002
Mallya M, Campbell RD, Aguado B (2006) Characterization of the five novel Ly-6 superfamily members encoded in the MHC, and detection of cells expressing their potential ligands. Protein Sci 15(10):2244–2256. https://doi.org/10.1110/ps.062242606
Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ (2010) Target-enrichment strategies for next-generation sequencing. Nat Methods 7(2):111–118. https://doi.org/10.1038/NMETH.1419
Matsumoto M, Zhou Y, Matsuo S, Nakanishi H, Hirose K, Oura H, Arase S, Ishida-Yamamoto A, Bando Y, Izumi K, Kiyonari H, Oshima N, Nakayama R, Matsushima A, Hirota F, Mouri Y, Kuroda N, Sano S, Chaplin DD (2008) Targeted deletion of the murine corneodesmosin gene delineates its essential role in skin and hair physiology. Proc Natl Acad Sci 105(18):6720–6724. https://doi.org/10.1073/pnas.0709345105
Meijaard E, D’Huart J-P, Oliver W (2011) Family Suidae (pigs). In: Wilson DE, Mittermeier RA (eds) Handbook of the mammals of the world, vol 2. Lynx Edicions, Barcelona, pp 248–291
Meng XJ (2012) Emerging and re-emerging swine viruses. Transbound Emerg Dis 59:85–102. https://doi.org/10.1111/j.1865-1682.2011.01291.x
Meyer M, Kircher M (2010) Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc 2010(6). https://doi.org/10.1101/pdb.prot5448
Mokry M, Feitsma H, Nijman IJ, de Bruijn E, van der Zaag PJ, Guryev V, Cuppen E (2010) Accurate SNP and mutation detection by targeted custom microarray-based genomic enrichment of short-fragment sequencing libraries. Nucleic Acids Res 38(10):e116. https://doi.org/10.1093/nar/gkq072
Mooseker MS, Cheney RE (1995) Unconventional myosins. Annu Rev Cell Dev Biol 11(1):633–675. https://doi.org/10.1146/annurev.cb.11.110195.003221
Moutou KA, Koutsogiannouli EA, Stamatis C, Billinis C, Kalbe C, Scandura M, Mamuris Z (2013) Domestication does not narrow MHC diversity in Sus scrofa. Immunogenetics 65(3):195–209. https://doi.org/10.1007/s00251-012-0671-8
Mukherjee S, Sarkar-Roy N, Wagener DK, Majumder PP (2009) Signatures of natural selection are not uniform across genes of innate immune system, but purifying selection is the dominant signature. Proc Natl Acad Sci 106(17):7073–7078. https://doi.org/10.1073/pnas.0811357106
Na Ayudhya SN, Assavacheep P, Thanawongnuwech R (2012) One world-one health: the threat of emerging swine diseases. An Asian perspective. Transbound Emerg Dis 59:9–17. https://doi.org/10.1111/j.1865-1682.2011.01309.x
Nagata T, Weiss EH, Abe K, Kitagawa K, Ando A, Yara-Kikuti Y, Seldin MF, Ozato K, Inoko H, Taketo M (1995) Physical mapping of the retinoid X receptor B gene in mouse and human. Immunogenetics 41(2-3):83–90
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3(5):418–426
Nei M, Gu X, Sitnikova T (1997) Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci U S A 94(15):7799–7806. https://doi.org/10.1073/pnas.94.15.7799
Nijman IJ, Mokry M, van Boxtel R, Toonen P, de Bruijn E, Cuppen E (2010) Mutation discovery by targeted genomic enrichment of multiplexed barcoded samples. Nat Methods 7(11):913–915. https://doi.org/10.1038/nmeth.1516
Van Oosterhout C (2009) A new theory of MHC evolution: beyond selection on the immune genes. Proc Biol Sci 276(1657):657–665. https://doi.org/10.1098/rspb.2008.1299
Peñalba JV, Smith LL, Tonione MA, Sass C, Hykin SM, Skipwith PL, McGuire JA, Bowie RCK, Moritz C (2014) Sequence capture using PCR-generated probes: a cost-effective method of targeted high-throughput sequencing for nonmodel organisms. Mol Ecol Resour 14:1000–1010. https://doi.org/10.1111/1755-0998.12249
Penn DJ, Ilmonen P (2001) Major histocompatibility complex (MHC). In: Encyclopedia of life sciences. John Wiley & Sons, Ltd, Chichester, pp 1–7
Piertney SB, Oliver MK (2006) The evolutionary ecology of the major histocompatibility complex. Heredity (Edinb) 96(1):7–21. https://doi.org/10.1038/sj.hdy.6800724
Pink RC, Wicks K, Caley DP, Punch EK, Jacobs L, Raul D, Carter F (2011) Pseudogenes: pseudo-functional or key regulators in health and disease? RNA 17(5):792–798. https://doi.org/10.1261/rna.2658311.transcription
Piontkivska H, Rooney AP, Nei M (2002) Purifying selection and birth-and-death evolution in the histone H4 gene family. Mol Biol Evol 19(5):689–697. https://doi.org/10.1093/oxfordjournals.molbev.a004127
R Development Core Team (2008) Computational many-particle physics. Springer Berlin Heidelberg, Berlin
Renard C, Vaiman M, Chiannilkulchai N, Cattolico L, Robert C, Chardon P (2001) Sequence of the pig major histocompatibility region containing the classical class I genes. Immunogenetics 53(6):490–500. https://doi.org/10.1007/s002510100348
Renard C, Chardon P, Vaiman M (2003) The phylogenetic history of the MHC class I gene families in pig, including a fossil gene predating mammalian radiation. J Mol Evol 57(4):420–434. https://doi.org/10.1007/s00239-003-2491-9
Renard C, Hart E, Sehra H, Beasley H, Coggill P, Howe K, Harrow J, Gilbert J, Sims S, Rogers J, Ando A, Shigenari A, Shiina T, Inoko H, Chardon P, Beck S (2006) The genomic sequence and analysis of the swine major histocompatibility complex. Genomics 88(1):96–110. https://doi.org/10.1016/j.ygeno.2006.01.004
Rodgers JR, Cook RG (2005) MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol 5(6):459–471. https://doi.org/10.1038/nri1635
Rodríguez-Fernández JL, Cabañas LG (2013) Chemoattraction: basic concepts and role in the immune response. eLS. https://doi.org/10.1002/9780470015902.a0000507.pub3
Rohland N, Reich D (2012) Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res 22(5):939–946. https://doi.org/10.1101/gr.128124.111
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3):539–542. https://doi.org/10.1093/sysbio/sys029
Rosenbaum DM, Rasmussen SGF, Kobilka BK (2009) The structure and function of G-protein-coupled receptors. Nature 459(7245):356–363. https://doi.org/10.1038/nature08144
Sánchez-Torres C, Gómez-Puertas P, Gómez-Del-Moral M, Alonso F, Escribano JM, Ezquerra A, Domínguez J (2003) Expression of porcine CD163 on monocytes/macrophages correlates with permissiveness to African swine fever infection. Arch Virol 148(12):2307–2323. https://doi.org/10.1007/s00705-003-0188-4
Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W (2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res 10(4):577–586. https://doi.org/10.1101/gr.10.4.577
Shigenari A, Ando A, Renard C, Chardon P, Shiina T, Kulski JK, Yasue H, Inoko H (2004) Nucleotide sequencing analysis of the swine 433-kb genomic segment located between the non-classical and classical SLA class I gene clusters. Immunogenetics 55(10):695–705. https://doi.org/10.1007/s00251-003-0627-0
Simon M, Montézin M, Guerrin M, Durieux JJ, Serre G (1997) Characterization and purification of human corneodesmosin, an epidermal basic glycoprotein associated with corneocyte-specific modified desmosomes. J Biol Chem 272(50):31770–31776. https://doi.org/10.1074/jbc.272.50.31770
Sommer S (2005) The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front Zool 2(1):16. https://doi.org/10.1186/1742-9994-2-16
Spurgin LG, Richardson DS (2010) How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc Biol Sci 277(1684):979–988. https://doi.org/10.1098/rspb.2009.2084
Stam M, Hayes H, Bertaud M, Teillaud A, Lemonnier G, Rogel-Gaillard C (2008) Centromeric/pericentromeric junction within the MHC locus on chromosome 7 in pig. In: XXXI Conference of the International Society for Animal Genetics, Amsterdam, Netherlands
Taber A, Altrichter M, Beck H, Gongora J (2011) Family Tayassuidae (peccaries). In: Wilson DE, Mittermeier RA (eds) Handbook of the mammals of the world, vol 2. Lynx Edicions, Barcelona, pp 292–307
Takahashi K, Rooney AP, Nei M (2000) Origins and divergence times of mammalian class II MHC gene clusters. J Hered 91(3):198–204. https://doi.org/10.1093/jhered/91.3.198
Tanaka-Matsuda M, Ando A, Rogel-Gaillard C, Chardon P, Uenishi H (2009) Difference in number of loci of swine leukocyte antigen classical class I genes among haplotypes. Genomics 93(3):261–273. https://doi.org/10.1016/j.ygeno.2008.10.004
Tennant LM, Renard C, Chardon P, Powell PP (2007) Regulation of porcine classical and nonclassical MHC class I expression. Immunogenetics 59(5):377–389. https://doi.org/10.1007/s00251-007-0206-x
Treangen TJ, Salzberg SL (2011) Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 13(1):36–46. https://doi.org/10.1038/nrg3117
Wilming LG, Gilbert JGR, Howe K, Trevanion S, Hubbard T, Harrow JL (2007) The Vertebrate Genome Annotation (VEGA) database. Nucleic Acids Res 36(Database):D753–D760. https://doi.org/10.1093/nar/gkm987
Xia X (2017) DAMBE6: new tools for microbial genomics, phylogenetics, and molecular evolution. J Hered 108(4):431–437. https://doi.org/10.1093/jhered/esx033
Yeager M, Hughes AL (1999) Evolution of the mammalian MHC: natural selection, recombination, and convergent evolution. Immunol Rev 167(1):45–58. https://doi.org/10.1111/j.1600-065X.1999.tb01381.x
Zozulya S, Echeverri F, Nguyen T (2001) The human olfactory receptor repertoire. Genome Biol 2(6):research0018.1–research001812. https://doi.org/10.1186/gb-2001-2-6-research0018
Acknowledgements
We thank the Sydney School of Veterinary Science at the University of Sydney for providing research funding for sampling and financial support, allowing J. Gongora to undertake preliminary experiments in Australia and a sabbatical period to generate data for this project at INRA. We thank INRA for making funding available from The French National Research Agency (PSC-08-GENO-CapSeqAn). All samples were provided by J. Gongora, collected by himself or accessed through Dr. Stewart Lowden, Dr. Joeke Nijboer (Rotterdam Zoo) or through collaboration with institutions in Eurasia, Africa and the Americas. Many thanks to Bertrand Bed’hom for the constructive feedback on the manuscript and for the advice on the MHC locus, and the INRA @BRIDGe platform where the hybridisation capture experiments were performed.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, C., Moroldo, M., Perdomo-Sabogal, A. et al. Inferring the evolution of the major histocompatibility complex of wild pigs and peccaries using hybridisation DNA capture-based sequencing. Immunogenetics 70, 401–417 (2018). https://doi.org/10.1007/s00251-017-1048-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-017-1048-9