Abstract
Research on microbial communities associated with wild animals provides a valuable reservoir of knowledge that could be used for enhancing their rehabilitation and conservation. The loggerhead sea turtle (Caretta caretta) is a globally distributed species with its Mediterranean population categorized as least concern according to the IUCN Red List of Threatened Species as a result of robust conservation efforts. In our study, we aimed to further understand their biology in relation to their associated microorganisms. We investigated epi- and endozoic bacterial and endozoic fungal communities of cloaca, oral mucosa, carapace biofilm. Samples obtained from 18 juvenile, subadult, and adult turtles as well as 8 respective enclosures, over a 3-year period, were analysed by amplicon sequencing of 16S rRNA gene and ITS2 region of nuclear ribosomal gene. Our results reveal a trend of decreasing diversity of distal gut bacterial communities with the age of turtles. Notably, Tenacibaculum species show higher relative abundance in juveniles than in adults. Differential abundances of taxa identified as Tenacibaculum, Moraxellaceae, Cardiobacteriaceae, and Campylobacter were observed in both cloacal and oral samples in addition to having distinct microbial compositions with Halioglobus taxa present only in oral samples. Fungal communities in loggerheads’ cloaca were diverse and varied significantly among individuals, differing from those of tank water. Our findings expand the known microbial diversity repertoire of loggerhead turtles, highlighting interesting taxa specific to individual body sites. This study provides a comprehensive view of the loggerhead sea turtle bacterial microbiota and marks the first report of distal gut fungal communities that contributes to establishing a baseline understanding of loggerhead sea turtle holobiont.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microbial communities associated with animal hosts play crucial roles in various aspects of the host’s development, physiology, immune response, metabolism, and reproduction, and may have an impact on the host’s evolutionary potential [1, 2]. While the importance of sea turtle epibiosis with macro-epibionts (> 1 mm) such as barnacles has long been recognized [3], research on the microbial epibionts and endobionts of sea turtles has only recently gained attention [4,5,6]. Sea turtles hold a unique ecological role as keystone species, connecting terrestrial and coastal habitats, but they are also highly vulnerable to anthropogenic threats, such as climate change, disruption of feeding and breeding habitats, egg poaching, and accidental bycatch [7,8,9]. To lessen some of the pressure sea turtles face today, global conservation efforts have focused on safeguarding female turtles and their nesting areas, together with the rehabilitation of injured and sick turtles [8]. Motivated by the aim of enhancing the rehabilitation and conservation of wild animals and their associated microbiomes, the studies of sea turtle gut, skin, egg, and nest microbiomes have become a forefront in sea turtle conservation research, building upon cultivation-based and pathogen-oriented research in the veterinary domain [10,11,12].
Loggerhead sea turtles (Caretta caretta) are a widely distributed species and are classified as vulnerable by the IUCN Red List of threatened species [13]. However, the Mediterranean subpopulation of loggerheads is categorized as “Least Concern” due to long-term successful conservation efforts [8, 13]. Currently, loggerhead sea turtles’ microbiota is the second most studied, preceded only by green turtles (Chelonia mydas) [6]. Previous studies that used culture-dependent approaches have identified the most common pathogens associated with mucosal surfaces, skin lesions (such as bacterial families Aeromonadaceae, Pseudomonadaceae, Enterobacteriaceae), and hatchling failure (Fusarium spp.), with the presence of antibiotic resistance genes indicating loggerheads as sentinels of antibiotic pollution in the Mediterranean [11, 14,15,16,17,18,19,20,21,22]. Recent investigations using next-generation sequencing approaches to study the loggerhead microbiota shed light on the bacterial community structure and composition of the gastrointestinal tract [5, 23, 24], the impact of rehabilitation on mucosal bacteriomes [24, 25], the effects of plastic pollution on the gut bacteria [26], variations in microbial communities driven by localities [27, 28] or turtle anatomy [28, 29], and host-microbial coevolution patterns [30]. On the other hand, the fungal communities associated with marine reptiles, including sea turtles, have received limited attention using cultivation-independent approaches, despite the vulnerability of sea turtles to infections caused by Fusarium spp. during early development [18]. Recent work by Guo et al. [31] provided initial insights into the fungal communities found on carapace (healthy and ulcerated), in faeces, and in the seawater of green turtle juveniles undergoing rehabilitation; however, comprehensive surveys of endobiotic fungal communities in loggerhead sea turtles have not yet been conducted. Given the ecological significance of loggerhead sea turtles in the Mediterranean basin ecosystem, their role as sentinels for pollution, and their potential to act as vectors for zoonotic diseases, a comprehensive approach including eukaryotic microorganisms is necessary to understand the loggerhead sea turtle microbiota. This knowledge will contribute to the advancement of current conservation practices and future microbial stewardship efforts [32].
The objective of this study was to investigate the bacterial and fungal communities associated with loggerhead sea turtles found in the Adriatic Sea. More specifically, we aimed to analyse the bacterial communities in the cloacal, oral, and enclosure samples, as well as fungal communities of cloacal and enclosure tank water samples using amplicon sequencing targeting the V3-V4 (V34) region of 16S rRNA gene and the ITS2 region of nuclear ribosomal genes, respectively. Furthermore, when available, we aimed to compare the bacterial communities of carapace biofilm samples corresponding to turtles in this study out of which some were previously analysed as a part of our earlier study on sea turtle epibiosis [28]. By combining these datasets, we provide a comprehensive overview of the environmental, surface, and internal microbiota of the loggerhead turtles, establishing a baseline for future holobiont approaches to studying the loggerhead sea turtles.
Methods
Loggerhead Sea Turtle Sampling
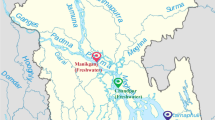
Loggerhead sea turtles investigated in this study were found at various locations along the Adriatic Sea coast from 2019 to 2021 (Fig. 1a) and transported to two locations where the sampling was conducted: at the Sea Turtle Clinic (STC) of the Department of Veterinary Medicine of University of Bari “Aldo Moro” in Italy and the Sea Turtle Rescue Center Aquarium Pula in Croatia. The turtles were sampled immediately upon their arrival to the rehabilitation centres or during/after rehabilitation, and prior to release (Table S1). A total of 18 loggerhead sea turtles and 8 respective enclosures were included in the sampling. Turtles were classified as juveniles (n = 10), subadult (n = 4), and adults (n = 4) according to their size [33] and sex determination was based on observable physical characteristics when possible (Table 1; Fig. 1b). Additional information about sampling procedures and the loggerhead population surveyed in this study can be found in Supplementary Methods and Table S2.
Locations and body measurements of loggerhead sea turtles with corresponding IDs. (a) Map of locations where loggerhead sea turtles were found prior to transport to rehabilitation centres and sampling. (b) Relationship of loggerheads’ weight in kilogrammes and curved carapace length in centimetres (CCL). The sex of each turtle is indicated by shape [female triangle, male square, and not determined (ND) circle], while age range was determined as follows: juveniles ≤ 59.9 cm, subadults 60–69.9 cm, adults ≥ 70 cm
The endozoic samples were collected from cloacal and oral cavities in triplicate by sterile synthetic swabs (Aptaca Nuova) as described in Filek et al. [25]. When available, enclosure tank water was collected in sterile containers, vacuum filtered on 0.2-μm sterile Whatman polycarbonate membrane filters (Sigma-Aldrich), and stored in 2-ml tubes in 96% EtOH. All samples were stored at −20 °C until DNA extraction and further processing. Additionally, corresponding epizoic carapace biofilm samples were obtained by randomly brushing the entire carapace using a toothbrush (Dentalux Classic, hard, Lidl) according to [28, 34] and the collected material was resuspended in 96% EtOH, and stored at −20 °C (Table 1). Each endozoic sequencing sample ID that is referred to in this manuscript has a 16S or ITS prefix, sampling event number, and suffix corresponding to sampling site: C — cloaca, O — oral cavity, W — tank water; for example, sample ID ITS0084C represents cloacal sample of fungal ITS2 sequences for sampling event 0084 (turtle ID010; Table 1). Epizoic samples have a TB prefix and numbering unrelated to sampling event.
DNA Extraction and Sequencing
Total DNA from swabs and filters was extracted using the DNeasy PowerSoil kit (Qiagen) following the manufacturer’s instructions with several modifications: (1) the samples were incubated in C1 solution at 65 °C for 1 h; (2) instead of bead beating, PowerBead Tubes were vortexed horizontally for 10 min at maximum speed; and (3) all downstream incubation times at 2–8 °C were increased to 15 min.
The methods used for epizoic carapace biofilm samples that were sequenced for V4 region (ID010–ID074) were described by Kanjer et al. [28]. The DNA from biofilm scrapings (ID093–ID122) analysed only in this study was extracted via DNeasy PowerLyzer PowerSoil extraction kit (Qiagen) following the manufacturer’s instructions and modified as follows: (1) after addition of C1 solution, the samples were incubated at 70 °C for 10 min; (2) bead beating was performed at 30 Hz for 1 min in TissueLyzer Retsch Qiagen; (3) 50 µl of C6 solution was used for DNA elution and incubated for 5 min at room temperature prior to centrifugation. Nuclease-free water (W4502, Sigma-Aldrich) was used as the negative control for the DNA extraction step and was processed using the DNA extraction kit in parallel to all the samples. The quality and quantity of extracted DNA were evaluated by BioSpec-nano (Shimadzu).
The extracted DNA from both endozoic and epizoic samples was stored at −20 °C and sent for Illumina MiSeq v3 300 × 2 bp paired-end sequencing to Microsynth, Switzerland. Primers used for sequencing the V34 region of 16S rRNA gene were 341F and 805R [35], and primers for fungal ITS2 region of the nuclear ribosomal gene were ITS3 and ITS4 [36].
Bioinformatics and Statistics
The obtained sequences had non-biological sequences trimmed by the sequencing facility and checked for quality with FastQC [37]. Sequencing data is available at EMBL ENA at accessions PRJEB62752 and PRJEB68298 for 16S rRNA gene, PRJEB62762 for ITS2 region sequences, and from Kanjer et al. [28] PRJEB51458. The sequences were imported and analysed in QIIME 2 (versions 2021.8 and 2023.2) [38].
Statistical analyses were performed within QIIME 2 environment and with R. Alpha diversity indices, including Shannon’s entropy, Pielou’s evenness, Faith’s phylogenetic diversity, and observed ASVs, were calculated via q2-diversity plugin. The Kruskal-Wallis rank-sum test was employed to determine differences between selected groups, followed by post hoc pairwise comparisons using the Wilcoxon rank-sum exact test. For beta diversity analyses, rarefied data (sequencing depth determined by alpha rarefaction curves) were explored using the q2-diversity plugin with Bray-Curtis, Jaccard, unweighted UniFrac, and weighted UniFrac distances [39]. Compositional data analysis on non-rarefied datasets was performed by calculating robust Aitchison distances using the q2-deicode plugin or the R package vegan v.2.6-4 [40,41,42]. Principal coordinate analysis (PCoA) was conducted on Bray-Curtis, Jaccard, and all UniFrac distances, while principal component analysis was performed for robust Aitchison (rPCA) using q2-diversity and q2-deicode, respectively. To assess the relative impact of factors (age range, sex, and duration of rehabilitation) on microbial communities, a multi-way permutational multivariate analysis of variance (Adonis2 PERMANOVA) with 9999 permutations was employed (Anderson, 2001) in R by using vegan v.2.6-4 and pairwise Adonis v.0.4.1 packages [43]. Resulting p-values from all pairwise tests were adjusted using the Benjamini-Hochberg false-discovery rate (FDR) correction for multiple comparisons (reported as q-values). Differential abundance analysis was used to identify differentially abundant (DA) features in sample site pairs by using the ANCOMBC package “ancombc2” function in R [44, 45]. Default parameters were used and pairwise testing was enabled (with Holm’s method for adjusting p-values, reported as q-values), except for DA testing in V4-trimmed sequences where the “struc_zero” was set to “TRUE” to exclude structural zeros based on sampling sites. Log fold change (LFC) indicates the scale of differential abundance between differentially abundant features. Features with p-value < 0.05 were reported as differentially abundant, and q-value < 0.05 as significantly differentially abundant.
Data exploration and visualizations were conducted by using R v.4.3.0 in RStudio (R Core Team 2023) with packages listed above and in Supplement and qiime2R v.0.99.6 [46], tidyverse v2.0.0 [47], ggplot2 v3.4.2 [48], Microsoft Excel, and Adobe Illustrator. Additional details of sample processing and data analyses are available in the Supplementary Methods.
Results
Endozoic and Tank Water Bacterial Communities
Altogether, 50 endozoic and water samples (plus one negative control) were sequenced, and 7,110,067 high-quality sequences were obtained (median frequency per sample was 107,522, min. 2, max. 911,443). Denoising yielded a total of 11,105 ASVs. After filtering mitochondrial and chloroplast sequences, the number of ASVs decreased to 10,946. Forty-three samples yielded enough high-quality reads (at minimum sequencing depth above 10,000) for downstream analyses (19/21 cloacal, 16/20 oral, and 8/9 tank water samples).
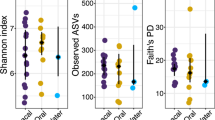
The highest number of observed features (OF) and phylogenetic diversity (Faith’s PD) was found in tank water samples (median OF = 603, IQR = 450; median Faith’s PD = 42.8, IQR = 53.9), followed by oral samples (median OF 473, IQR = 428; median Faith’s PD 40.2, IQR = 26.2), and cloacal samples (median OF = 308, IQR = 235; median Faith’s PD = 28.4, IQR = 20) (Fig. 2a). ASV richness and evenness (Shannon’s index) were highest in oral samples (median = 6.28, IQR = 1.33), followed by cloacal (median = 5.62, IQR = 1.66) and tank water samples (median = 5.44, IQR = 1.67) (Fig. 2b). Alpha diversity showed significant differences among sample sites (Kruskal-Wallis rank-sum test) for OF and Faith’s PD (p-value < 0.01), while pairwise sample site comparisons (Wilcoxon rank sum test) showed differences between cloaca vs. oral samples and cloaca vs. tank water (q-value < 0.05). Shannon’s index showed weak statistical difference among samples sites (p-value = 0.04). Pearson’s correlation on alpha diversity indices against size of the turtle (CCL) in individual sample site groups showed strong negative correlation between cloacal samples and OF and Faith’s PD (R = −0.67, p-value = 0.002 and R = −0.59, p-value = 0.007, respectively) (Fig. 2c), while Shannon’s index showed weaker negative correlation with CCL (R = −0.53, p-value = 0.021). Oral and tank water alpha diversity did not show any correlation effects. Further, within cloacal samples, differences were detected (Kruskal-Wallis p-value < 0.05) in age range (OF, pairwise Wilcox: adult vs. juvenile q-value = 0.024; Faith’s PD, pairwise: adult vs. juvenile q-value = 0.024) and sex of the turtles (OF, pairwise Wilcox: male vs. ND q-value = 0.004; Faith’s PD, pairwise: male vs. ND q-value = 0.018), which are directly related to turtles’ size.
Bacterial community structure and diversity in loggerhead sea turtles’ cloacal, oral, and tank water samples. Alpha diversity boxplots with sample density for Faith’s phylogenetic diversity (a) and Shannon’s index (b) per sample site. (c) Pearson’s correlation between Faith’s phylogenetic diversity and curved carapace length (CCL). (d) Robust Aitchison PCA biplot with highly ranked features as loadings. Cloacal samples are depicted by diamonds, oral samples by circles, and tank water by inverted triangle shapes
Bacterial communities among samples sites differed based on PERMANOVA for Bray-Curtis (R2 = 0.09, Pr(> F) = 0.001), Jaccard (R2 = 0.08, Pr(> F) = 0.001), unweighted and weighted UniFrac (R2 = 0.12, Pr(> F) = 0.001; R2 = 0.13, Pr(> F) = 0.001, respectively), and robust Aitchison distances (R2 = 0.24, Pr(> F) = 0.002). Pairwise PERMANOVA detected differences between all sample site pairs for most beta diversity metrics except robust Aitchison where only cloaca vs. tank water and tank water vs. oral samples pairs were significantly different (Pr(> F) ≤ 0.002). Highly ranked ASVs impacting the distribution of samples on rPCA biplot were assigned to Gammaproteobacteria, order Oceanospirillales, Shewanella algae, Vibrio sp., and NS3a marine group (Fig. 2d). Within cloacal samples, significant differences in bacterial communities were detected between adults vs. juveniles (Bray-Curtis, Jaccard, and unweighted UniFrac Pr(> F) < 0.05) and ND vs. males (all distances except robust Aitchison Pr(> F) < 0.05) or females (Jaccard and weighted UniFrac Pr(> F) < 0.05). In oral samples, there were differences between early vs. late hospitalization duration (all distances except robust Aitchison Pr(> F) < 0.05) and early vs. mid (weighted UniFrac Pr(> F) < 0.05) hospitalization durations.
Cloacal bacterial communities differed between age groups based on PERMANOVA for Bray-Curtis (R2 = 0.15, Pr(> F) = 0.018), Jaccard (R2 = 0.14, Pr(> F) = 0.008), and unweighted UniFrac distance (R2 = 0.16, Pr(> F) = 0.013). Pairwise PERMANOVA showed significant differences only between adults and juveniles for all above-mentioned diversity measures (Pr(> F) ≤ 0.009). The differences between adults and juveniles are observed in community composition and structure as well: phylum Verrucomicrobiota appears more often in juveniles, juveniles also have higher RA of Rhodobacterales (Alphaproteobacteria), Cardiobacterales (Gammaproteobacteria), Kineosporiales (Actinobacteriota), Oligoflexales (Bdellovibrionota), Arcobacteraceae (Campylobacterales), and more Flavobacteriales (Bacteroidota) than adults. On the other hand, adults carry more Bacteroidales (Bacteroidota), Pasteurellales (Gammaproteobacteria), Helicobacteraceae and Campylobacteraceae (Campylobacterales), and Leptotrichiaceae (Fusobacteriales). The low number of samples (≤ 10) in each age group prevented us from further differential abundance analyses. Detailed taxonomic composition of bacterial communities found in oral, cloacal, and tank water samples is reported in Supplementary Results, Table S3 with additional visualizations available at Github.
Differential abundance analysis detected 33 significantly DA features (post adjusting for multiple testing) out of which two were differentially abundant in cloaca (ASV467 Rhodobacteraceae and ASV4007 Shewanella algae), and three in oral samples (ASV9794 Truepera sp., ASV6166 and ASV467 both belonging to Rhodobacteraceae) (Fig. 3a). Tank water had 28 DA ASVs when tested against cloaca or oral samples, most of which belonged to typical marine taxa (Cryomorphaceae, NS3a marine group, SAR406 clade, Rhodobacteraceae, Nitrincolaceae, etc.). When collapsed to species level, 75 significantly DA taxa were detected, out of which 13 in cloaca and 15 in oral samples (Fig. 3b). Depending on the tested sample site pair, several features would be DA in both oral and cloacal samples when compared to tank water, e.g., genera Marinifilum, Tenacibaculum, and Labrenzia, members of Cardiobacteriaceae and Comamonadaceae families (Fig. 3b).
Differential abundance analysis by ANCOM-BC2 for ASVs (a) and taxa collapsed to species level (b) in cloacal, oral, and tank water samples. Only differentially abundant taxa in oral or cloacal sample sites are shown. Sample site abbreviations are “Clo” for cloaca; “Orl” for oral; and “TW” for tank water samples. The first sample site listed in sample site pairs was used as a denominator for log fold change (LFC) calculations in pairwise testing, thus the LFC value for that body site being < 0. Single asterisk (*) indicates significant differential abundance (p-value < 0.05), while double asterisk (**) indicates significance after adjusting for multiple testing (q-value < 0.05)
Endozoic and Tank Water Fungal Communities
Overall, 29 samples (plus one negative control) were sequenced, and 2,208,729 high-quality sequences were obtained (median frequency per sample was 69,203, min. 11,722, max. 150,142). Denoising yielded a total of 9547 ASVs. Based on alpha rarefaction curves, 27 samples yielded enough high-quality reads (at minimum sequencing depth at 25,000 reads) for downstream statistical analyses requiring rarefied data (19/20 cloacal and 8/9 tank water samples). All samples except negative control were used in compositional data analyses.
Tank water samples had significantly higher number of observed features and phylogenetic diversity (median = 674, IQR = 97.8; Faith’s PD median = 98.0, IQR = 10.5) than cloacal samples (median = 509, IQR = 372, Faith’s PD median = 66.6, IQR = 51.7) (Fig. 4a) based on the Kruskal-Wallis rank-sum test (OF p-value = 0.008; Faith’s PD p-value = 0.007). Shannon’s diversity was not detected as significantly different between sample sites (tank water median = 8.66, IQR = 1.31; cloaca median = 6.76, IQR = 2.58) (Fig. 4b). There were no significant differences in alpha diversity values within cloacal or tank water fungal communities when tested for age range, sex, or rehabilitation duration. The only beta diversity metric that showed significant differences between tank water and cloaca was unweighted UniFrac (R2 = 0.06, Pr(> F) = 0.042). Unweighted UniFrac PCoA showed unclear sample groupings; however, there is a separation of cloacal and corresponding tank water samples on PCA1 axis (Fig. S1a, corresponding samples connected with dashed lines). Compositional data analysis rPCA did not show clear separation of samples sites, and top ranked ASVs belonged to Preussia flanaganii, genus Tetracladium, order Xylariales, genus Rhizoctonia, and family Nectriaceae (Fig. S1b). When collapsed to species level, cloacal and tank water samples each had five differentially abundant fungal taxa detected. In cloacal samples, DA taxa were Leptodiscella, Elaphomyces, Sclerocleista, Gliomastix, and Thelephora; and in tank water, they were Cladosporium, unidentified Leotiomycetes, unidentified Sordariomycetes (Chaetosphaerilaes), Nectria, and Humicola — although none of them was statistically significant after correction for multiple testing (Fig. 4c). Cloacal and tank water fungal communities were represented mostly by phyla Ascomycota (average RA ± standard deviation in cloaca and tank water = 57 ± 15% and 53 ± 9%, respectively), Basidiomycota (19 ± 9% and 22 ± 3%), Glomeromycota (7 ± 6% and 10 ± 12%), Mortierellomycota (2 ± 2% and 3 ± 2%), and unidentified Fungi (13 ± 20% and 11 ± 7%) (Fig. 4d). Detailed taxonomic composition of fungal communities found in cloacal and tank water samples is reported in Supplementary Results and Table S4.
Structure and composition of fungal microbial communities in cloaca and tank water samples. Alpha diversity boxplots with sample density for Faith’s phylogenetic diversity (a) and Shannon’s index (b) per sample site. (c) Differential abundance analysis by ANCOM-BC2 for taxa collapsed to species level in cloacal and tank water samples. Cloacal sample site was used as a denominator for log fold change (LFC) calculations in pairwise testing, thus the cloacal LFC values being < 0. Single asterisk (*) indicates significant differential abundance (p-value < 0.05). (d) Relative abundance of fungal phyla in cloacal and tank water samples present above 1% in at least one sample. Sample IDs ending with “C” belong to cloacal samples, while sample IDs ending with “W” belong to tank water samples
Epizoic and Endozoic Bacterial Communities
After trimming epizoic, endozoic, and tank water sequences to V4 region of 16S rRNA gene (65 samples in total) and denoising, 10,072,866 high-quality sequences were merged across all sequencing events (median frequency per sample was 126,629, min. 0, max. 946,183) and yielded 13,263 ASVs. Due to reduced resolution after trimming the longer V34 region to shorter V4 region, the number of ASVs for cloacal, oral, and tank water samples was expectedly lower (7725 in V4 vs. 11,105 in V34 sequences).
Carapace samples had the highest median species richness and diversity (OF median 732, IQR = 420, Faith’s PD median = 45.7, IQR = 25.2; median Shannon’s index = 5.94, IQR = 2.22), which was followed by tank water, oral, and cloacal samples (Fig. 5a and b). The differences between sample sites’ alpha diversity were detected as statistically significant (Kruskal-Wallis rank-sum test); however, pairwise comparisons showed differences only in Faith’s phylogenetic diversity and OF: between cloaca and all other sample sites (except tank water based on OF), oral cavity and carapace. All beta diversity metrics showed significant differences between body sites for Bray-Curtis (R2 = 0.13, Pr(> F) = 0.001), Jaccard (R2 = 0.10, Pr(> F) = 0.001), unweighted and weighted UniFrac (R2 = 0.16, Pr(> F) = 0.001; R2 = 0.18, Pr(> F) = 0.001, respectively), and robust Aitchison distances (R2 = 0.09, Pr(> F) = 0.001). Pairwise PERMANOVA detected differences between all sample site pairs (Pr(> F) < 0.01). Highly ranked ASVs impacting the distribution of samples on rPCA biplot belonged to Pseudoalteromonas sp., Vibrio sp., Shewanella sp., uncultured Saccharospirillaceae, uncultured Marinifilum, and uncultured Cardiobacteriaceae (Fig. 5c). The taxonomic composition of endozoic samples and tank water using the V4 sequences resembled the one detected with V34 and it is reported in Table S5. Additional information on taxonomic composition of epizoic bacterial communities in carapace samples is reported in Supplementary Results.
Bacterial community structure and diversity in loggerhead sea turtles’ carapace, cloacal, oral, and tank water samples. Alpha diversity boxplots with sample density for Faith’s phylogenetic diversity (a) and Shannon’s index (b) per sample site. (c) Robust Aitchison PCA biplot with highly ranked features as loadings. Cloacal samples are depicted by diamonds, oral samples by circles, carapace samples by squares, and tank water by inverted triangle shapes
Differential abundance analysis on ASVs (structural zeros excluded) detected 17 significantly DA ASVs across all sample sites (Fig. 6). A member of Rhodobacteraceae family (v4ASV8861) and Halioglobus sp. (v4ASV11062) were consistently DA in oral samples while the same goes for Cardiobacterium sp. (v4ASV10814) and a member of Enterobacteriaceae family (v4ASV11383) in cloaca. In carapace samples, a member of Flavobacteriaceae family (v4ASV2446), cyanobacteria Leptolyngbya sp. (v4ASV4472), Ahrensia sp. (v4ASV8480), Sphingomonadaceae (v4ASV9910), Erythrobacter sp. (v4ASV9994), Gammaproteobacteria (v4ASV10111), Alteromonas sp. (v4ASV10327), Arenicella sp. (v4ASV10634), Psychrobacter sp. (v4ASV12017), and Vibrio sp. (v4ASV12307) were DA relative to other sample sites, while Marinomonas sp. (v4ASV11704) was DA abundant in carapace tested against endozoic samples, but not tank water. In tank water, a member of Flavobacteriaceae (v4ASV2446) and NS3a marine group (v4ASV2817) were consistently DA (Fig. 6). Differential abundance analysis on taxa collapsed to species level is reported in Supplementary Results and Fig. S2.
Differential abundance analysis by ANCOM-BC2 (with structural zeros excluded) for ASVs in carapace, cloacal, oral, and tank water samples. Sample site abbreviations are “Car” for carapace; “Clo” for cloaca; “Orl” for oral; and “TW” for tank water samples. The first sample site listed in sample site pairs was used as a denominator for log fold change (LFC) calculations in pairwise testing, thus the LFC value for that body site being < 0. Single asterisk (*) indicates differential abundance (p-value < 0.05), while double asterisk (**) indicates significance after adjusting for multiple testing (q-value < 0.05)
Discussion
This study provides an overview of the bacterial and fungal communities inhabiting oral and cloacal environments of loggerhead sea turtles. Additionally, we aimed to investigate bacterial communities in carapace samples in relation to the gastrointestinal tract and tank water to explore possible connections between two habitats. The carapace exhibited the highest bacterial diversity, followed by oral samples, influenced by the tank water environment, and then cloacal samples. Each sampling site had distinct microbial communities and cloacal bacterial diversity negatively correlated with turtle size and age. Conversely, fungal communities in the cloaca were distinct from tank water and showed high heterogeneity among individual turtles, with no discernible patterns related to age or sex.
Cloacal Bacterial Diversity and Structure Changes with the Turtle Age
Similarly to previous studies on cloacal microbiota in Adriatic loggerhead sea turtles, we observed changes in bacterial richness, diversity, and structure, negatively correlating with loggerheads’ CCL and, consequently, their age [24]. According to research on vertebrate gut microbiomes, it is expected that the bacterial diversity increases with body size in animals with complex digestive systems, like ruminants, and decreases in animals with simple guts, often omnivores or carnivores [49]. Loggerhead sea turtles are omnivores and exhibit an ontogenetic shift from oceanic-pelagic habitats as juveniles to neritic-benthic feeding grounds as adults. However, in the Mediterranean Sea, they employ an “amphi-habitat strategy”, where juveniles, subadults, and adults share feeding grounds and similar feeding behaviours [50, 51]. Differences in diet, jaw size, bite force, and diving ability between juveniles and adults may result in distinct diets and digestive physiologies, potentially reflected in cloacal bacterial communities as increased richness. Studies on loggerhead diet show no significant differences in stomach contents among adults, subadults, and juveniles, but it is worth noting that softer prey like tunicates or jellyfish may not be as detectable due to easier digestion and lack of hard remains in morphology-based studies [50].
In this study, juveniles exhibited higher abundance and more frequent presence of Flavobacteriaceae and Tenacibaculum spp. in oral and cloacal samples than subadult and adult turtles. Tenacibaculum spp. are marine pathogens possibly carried by cnidarians and ctenophores as vectors [52, 53], suggesting a higher proportion of soft prey in juveniles’ diet. This study was conducted during non-winter months when jellyfish populations increase due to higher water temperatures potentially influencing prey availability. Adults likely have access to these prey types but also consume more challenging prey inaccessible to juveniles. Notably, in other studies, Tenacibaculum spp. were highly abundant on loggerhead skin as well [28], indicating their propensity to inhabit marine vertebrate surfaces and gastrointestinal tracts, irrespective of animal’s diet. While in previous research the Mogibacteraceae family detected in faeces correlated with CCL [24], we did not detect it in our data, possibly due to different taxonomy databases being utilized (Greengenes vs. SILVA in our study). BLAST analysis showed ASVs assigned as Peptostreptococcales-Tissieralles closely related to Mogibacterium kristiansenii but it still did not exhibit a CCL correlation.
Loggerhead Cloacal Communities Are a Promising Source of Novel Campilobacterota
Members of the Campilobacterota phyla, particularly Arcobacteriaceae, Campylobacteraceae, and Helicobacteraceae, have recently been found to form unique, cold-adapted communities in ectothermic reptiles [54]. As expected, and based on their physiology and lifestyle, the Arcobacter genus, which can thrive at atmospheric oxygen levels and prefers lower temperatures, was present in all sample sites. However, it was more abundant in oral than in cloacal samples and was rarely detected in tank water. The Helicobacter and Campylobacter genera, which are vertebrate-associated, were observed in cloacal and occasionally oral samples. Notably, one adult male (ID057) showed a higher prevalence of Helicobacter, reaching up to 20% relative abundance in the cloaca, represented by a single ASV assigned as Helicobacter sp. and not found in any other sample. This suggests a potential overgrowth or infection despite the relatively good clinical status of the individual observed at the time. Therefore, like other reptiles, loggerhead sea turtles could serve as a valuable source of previously undiscovered cold-adapted Campylobacter and Helicobacter species, as well as mucosal Arcobacter species.
Oral Bacterial Communities Harbour Distinct Taxa and Could Reflect Recent Diet
In oral samples, the genera Truepera and Halioglobus showed differential abundance. Trueperaceae, a relatively new family, includes one isolate Truepera radiovictrix, known for radiation resistance and thriving in extreme conditions like other Deinococci members [55]. Truepera was also a dominant part of the oral microbiota in splendid japalure lizards [56], but its role and functions in reptile oral microbiomes remain unclear. Tolerance to extreme environments and ability to use diverse carbon sources may allow Truepera spp. to outcompete other taxa on reptilian oral mucosa. The genus Halioglobus is typically found in seawater and marine sediments, and has been associated with dinoflagellate blooms and starving, green-lipped mussels [57, 58]. We have previously reported Halioglobus in oral samples of loggerhead sea turtles [25]. Halioglobus bacteria likely derive from loggerhead prey such as mussels or oysters rather than being an intrinsic property of sea turtle mucosal surfaces, given the limited information on host-associated Halioglobus. Similarly, the high relative abundance of the genus Exiguobacterium in oral and cloacal samples of one juvenile loggerhead (ID069) sampled upon admission may indicate recent feeding, as some Exiguobacterium species are commonly found on shrimp or algae [59,60,61].
Carapace Microbiota Is Distinct, yet Can Harbour Taxa Specific to Oral or Cloacal Microbiota
The carapace and skin of loggerhead sea turtles harbour diverse microbial biofilms, rich in prokaryotes, microeukaryotes, and macroeukaryotes like barnacles and algae [3, 28]. These surface microbial communities vary with the turtle’s geographical location, carapace condition, and the sampled anatomical site [28, 29]. They can be considered as microbial reservoirs and “diversity hotspots” in otherwise scarce environments [62, 63]. In our study, we found expectedly distinct microbial communities in the carapace, oral mucosa, and cloaca, although many microbial taxa were present across all body sites, including potential zoonotic pathogens primarily from the cloaca. Oral and cloacal samples shared several differentially abundant microbial taxa (Tenacibaculum, Cardiobacteraceae, Campylobacter), suggesting co-inhabitation of the gastrointestinal tract. The carapace microbiota differed significantly from tank water, with indication of transfer of certain taxa from carapaces to tank water or vice versa. Taxa like Alteromonadaceae and Colwelliaceae (Thalassotalea), differentially abundant on carapaces in this study, were prominent in enclosure tank water in prior studies that did not examine carapace bacterial communities [24], possibly originating from captive turtles’ carapaces. The implications of the interplay between microbes from different body sites on loggerhead sea turtles in their natural habitats and during captivity are not yet clear. However, it is essential to consider surface microbiota and potential opportunistic pathogens, especially when rehabilitating severely injured and possibly immunocompromised individuals.
Fungal Communities of Loggerhead Cloaca Are Highly Heterogeneous and Diverse
Fungal communities in loggerhead sea turtles’ cloaca and tank water exhibit high variability among individuals, with greater diversity observed in tank water. In this study, we could not attribute differences in the composition and structure of cloacal mycobiota to the turtles’ age, sex, or hospitalization status, possibly due to the limited sample size per sampling site and condition assessed. Conversely, captive juvenile green turtles displayed more consistent mycobiota richness and diversity across various sampling sites and health conditions, unaffected by environmental fungi [31]. The taxonomic composition of cloacal and tank water fungal phyla in our study aligns with previously reported marine fungi groups found in green sea turtle faeces [31], as well as marine algicolous fungi, sediments, and sponges [64]. Due to the lower resolution of the ITS2 gene marker and many unassigned fungal ASVs beyond the family level, this study offers just a general overview and serves as a foundation for further exploration of specific groups of interest.
We sporadically detected pathogenic fungal genera [18], with nine pathogenic genera found in both cloaca and tank water. Among these, Fusarium species (family Nectriaceae) are recognized sea turtle pathogens linked to reduced hatchling success [65]. Our study identified Fusarium oxysporum (a known pathogen), Fusarium neocosmosporellium, and Fusarium waltergamsii, not previously reported as sea turtle pathogens but related to known pathogenic species in the Fusarium solani species complex [18, 66]. Fusarium ASVs were relatively less abundant, except in one sample where they reached 8%. However, some ASVs assigned to the Nectriaceae genus may belong to Fusarium species as ITS region can be insufficient in detecting Fusarium species [67], potentially affecting our reported abundance of pathogenic fungi in cloacal and tank water samples.
The origin of these fungi — whether they are metabolically active and intrinsic to the sampled turtle population or introduced from the environment as spores or through food — is unclear. The impact of these fungi on the host, the interaction with the turtle’s immune system and physiology, and their potential host association remain unknown. Many fungal ASVs detected in this study belong to primarily terrestrial taxa, suggesting possible terrestrial sources during rehabilitation or a lack of marine fungal sequences in taxonomy databases. Additionally, numerous reads were assigned only as “Fungi”, indicating either undiscovered fungal taxa, lack of representative sequences, or possible turtle host origin.
Low Biomass of Samples Is a Potential Limitation to Interpreting Results
Microbial composition results should be interpreted with caution due to low biomass collected with swabs and fewer fungal cells compared to bacterial cells in vertebrate guts [68]. Negative control (sterile water) sequenced with bacterial primers showed a small number of reads that were also detected in low-read samples, indicating potential contamination during DNA extraction and sequencing specifically for samples with low biomass (e.g. kitome) [69, 70]. Surprisingly, the negative control sequenced with fungal primers had significantly more reads than the bacterial negative control, containing numerous ASVs also found in cloacal and tank water samples. Although the community structures of cloacal and tank water samples differed from the negative control, indicating genuine fungal communities rather than random contaminants, the high number of reads from seemingly low-biomass samples or the “empty” negative control raises concerns about our sample handling and sequencing approach. To our knowledge, no studies addressed fungal contamination in DNA extraction kits as they do for prokaryotes, making it challenging to pinpoint the exact source of this fungal DNA. For future studies, we suggest including additional negative control samples at various sampling and processing stages when investigating undescribed gut-associated fungal communities.
Conclusion
Loggerhead sea turtle-associated microbial communities are crucial for understanding the biology of these endangered reptiles and supporting conservation. In this study, we examined bacterial and fungal communities in juvenile, subadult, and adult loggerhead sea turtles. Our findings revealed distinct microbial communities in the carapace, cloaca, and oral mucosa, with characteristic taxa for each site and shared taxa between cloacal and oral samples (e.g. Tenacibaculum, Moraxellaceae, Cardiobacteriaceae, and Campylobacter). Cloacal bacterial communities exhibited decreasing diversity and changing composition with turtle age, likely due to shifts in diet as juveniles develop stronger bite force and diving capabilities. Microbial exchange with the environment appears to occur, particularly from turtles to tank water, especially from the carapace. Fungal communities in cloaca and tank water displayed high heterogeneity across individuals, with no age or clinical patterns, possibly due to limited samples and low fungal biomass in cloacal samples. Loggerhead sea turtles host complex microbial communities, including potential bacterial and fungal pathogens, which pose risks to handlers or the general public as changing turtle behaviour leads to increased human-turtle interactions. Despite growing research on loggerhead microbiomes, defining a healthy microbiome beyond bacteria remains challenging. Future research should prioritize establishing a description of healthy loggerhead microbiomes at different developmental stages (from eggs and hatchlings to juveniles and adults) to enhance conservation practices and explore potential probiotics and prebiotics for addressing their current and future needs.
Data Availability
Sequencing data with non-biological sequences removed is available at EMBL ENA at accessions PRJEB62752 and PRJEB68298 for 16S rRNA gene and PRJEB62762 for ITS2 region sequences. Epizoic samples’ sequencing data obtained from Kanjer et al. [28] and used in this study can be found at ENA under accession PRJEB51458. Complete code and instructions for processing of the sequencing data and subsequent statistical analyses, results, and data visualizations are available at Github (https://github.com/kl-fil/2023-Filek_et_al._TBIOME_project) and ZENODO data depository (https://doi.org/10.5281/zenodo.8054926).
Change history
26 June 2024
A Correction to this paper has been published: https://doi.org/10.1007/s00248-024-02405-z
References
McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ (2013) Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci 110:3229. https://doi.org/10.1073/pnas.1218525110
Henry LP, Bruijning M, Forsberg SKG, Ayroles JF (2021) The microbiome extends host evolutionary potential. Nat Commun 12:5141. https://doi.org/10.1038/s41467-021-25315-x
Robinson NJ, Pfaller JB (2022) Sea Turtle epibiosis: global patterns and knowledge gaps. Front Ecol Evol 10:844021. https://doi.org/10.3389/fevo.2022.844021
Robinson NJ, Majewska R, Lazo-Wasem EA, Nel R, Paladino FV, Rojas L, Zardus JD, Pinou T (2016) Epibiotic diatoms are universally present on all sea turtle species. PLoS ONE 11:e0157011–e0157011. https://doi.org/10.1371/journal.pone.0157011
Abdelrhman KFA, Bacci G, Mancusi C, Mengoni A, Serena F, Ugolini A (2016) A first insight into the gut microbiota of the Sea Turtle Caretta caretta. Front Microbiol 7:1–5. https://doi.org/10.3389/fmicb.2016.01060
Kuschke SG (2022) What lives on and in the sea turtle? A literature review of sea turtle bacterial microbiota. Anim Microbiome 4:52. https://doi.org/10.1186/s42523-022-00202-y
Stanford CB, Iverson JB, Rhodin AGJ, van Paul P, Mittermeier RA, Kuchling G, Berry KH, Bertolero A, Bjorndal KA, Blanck TEG, Buhlmann KA, Burke RL, Congdon JD, Diagne T, Edwards T, Eisemberg CC, Ennen JR, Forero-Medina G, Frankel M, Fritz U, Gallego-García N, Georges A, Gibbons JW, Gong S, Goode EV, Shi HT, Hoang H, Hofmeyr MD, Horne BD, Hudson R, Juvik JO, Kiester RA, Koval P, Le M, Lindeman PV, Lovich JE, Luiselli L, McCormack TEM, Meyer GA, Páez VP, Platt K, Platt SG, Pritchard PCH, Quinn HR, Roosenburg WM, Seminoff JA, Shaffer HB, Spencer R, Van Dyke JU, Vogt RC, Walde AD (2020) Turtles and tortoises are in trouble. Curr Biol 30:R721–R735. https://doi.org/10.1016/j.cub.2020.04.088
Mazaris AD, Schofield G, Gkazinou C, Almpanidou V, Hays GC (2017) Global sea turtle conservation successes. Sci Adv 3:e1600730–e1600730. https://doi.org/10.1126/sciadv.1600730
Mazaris AD, Dimitriadis C, Papazekou M, Schofield G, Doxa A, Chatzimentor A, Turkozan O, Katsanevakis S, Lioliou A, Abalo-Morla S, Aksissou M, Arcangeli A, Attard V, El Hili HA, Atzori F, Belda EJ, Ben Nakhla L, Berbash AA, Bjorndal KA, Broderick AC, Camiñas JA, Candan O, Cardona L, Cetkovic I, Dakik N, de Lucia GA, Dimitrakopoulos PG, Diryaq S, Favilli C, Fortuna CM, Fuller WJ, Gallon S, Hamza A, Jribi I, Ben Ismail M, Kamarianakis Y, Kaska Y, Korro K, Koutsoubas D, Lauriano G, Lazar B, March D, Marco A, Minotou C, Monsinjon JR, Naguib NM, Palialexis A, Piroli V, Sami K, Sönmez B, Sourbès L, Sözbilen D, Vandeperre F, Vignes P, Xanthakis M, Köpsel V, Peck MA (2023) Priorities for Mediterranean marine turtle conservation and management in the face of climate change. J Environ Manage 339:117805. https://doi.org/10.1016/j.jenvman.2023.117805
Hird SM (2017) Evolutionary Biology needs wild microbiomes. Front Microbiol 8:725. https://doi.org/10.3389/fmicb.2017.00725
Ebani VV (2023) Bacterial infections in Sea turtles. Vet Sci 10:333. https://doi.org/10.3390/vetsci10050333
Dallas JW, Warne RW (2023) Captivity and animal microbiomes: potential roles of Microbiota for Influencing Animal Conservation. Microb Ecol 85:820–838. https://doi.org/10.1007/s00248-022-01991-0
Casale P, Tucker AD (2017) Caretta caretta (amended version of 2015 assessment). The IUCN Red List of Threatened Species 2017: e.T3897A119333622. https://doi.org/10.2305/IUCN.UK.2017-2.RLTS.T3897A119333622.en. Accessed 8 May 2024
Blasi MF, Migliore L, Mattei D, Rotini A, Thaller MC, Alduina R (2020) Antibiotic resistance of Gram-negative Bacteria from Wild Captured Loggerhead Sea turtles. Antibiotics 9:162–162. https://doi.org/10.3390/antibiotics9040162
Pace A, Dipineto L, Fioretti A, Hochscheid S (2019) Loggerhead sea turtles as sentinels in the western Mediterranean: antibiotic resistance and environment-related modifications of Gram-negative bacteria. Mar Pollut Bull 149:110575–110575. https://doi.org/10.1016/j.marpolbul.2019.110575
Pace A, Rinaldi L, Ianniello D, Borrelli L, Cringoli G, Fioretti A, Hochscheid S, Dipineto L (2019) Gastrointestinal investigation of parasites and Enterobacteriaceae in loggerhead sea turtles from Italian coasts. BMC Vet Res 15:1–9. https://doi.org/10.1186/s12917-019-2113-4
Capri FC, Prazzi E, Casamento G, Gambino D, Cassata G, Alduina R (2023) Correlation between Microbial Community and Hatching failure in Loggerhead Sea Turtle Caretta caretta. Microb Ecol. https://doi.org/10.1007/s00248-023-02197-8
Gleason FH, Allerstorfer M, Lilje O (2020) Newly emerging diseases of marine turtles, especially sea turtle egg fusariosis (SEFT), caused by species in the Fusarium solani complex (FSSC). Mycology 11:184–194. https://doi.org/10.1080/21501203.2019.1710303
Trotta A, Cirilli M, Marinaro M, Bosak S, Diakoudi G, Ciccarelli S, Paci S, Buonavoglia D, Corrente M (2021) Detection of multi-drug resistance and AmpC β-lactamase/extended-spectrum β-lactamase genes in bacterial isolates of loggerhead sea turtles (Caretta caretta) from the Mediterranean Sea. Mar Pollut Bull 164:112015. https://doi.org/10.1016/j.marpolbul.2021.112015
Trotta A, Marinaro M, Sposato A, Galgano M, Ciccarelli S, Paci S, Corrente M (2021) Antimicrobial Resistance in Loggerhead Sea turtles (Caretta caretta): a comparison between clinical and commensal bacterial isolates. Animals 11:2435. https://doi.org/10.3390/ani11082435
Alduina R, Gambino D, Presentato A, Gentile A, Sucato A, Savoca D, Filippello S, Visconti G, Caracappa G, Vicari D, Arculeo M (2020) Is Caretta Caretta a Carrier of Antibiotic Resistance in the Mediterranean Sea? Antibiotics 9:116–116. https://doi.org/10.3390/antibiotics9030116
Gambino D, Persichetti MF, Gentile A, Arculeo M, Visconti G, Currò V, Caracappa G, Crucitti D, Piazza A, Mancianti F, Nardoni S, Vicari D, Caracappa S (2020) First data on microflora of loggerhead sea turtle (Caretta caretta) nests from the coastlines of Sicily. Biol Open 9:bio045252–bio045252. https://doi.org/10.1242/bio.045252
Arizza V, Vecchioni L, Caracappa S, Sciurba G, Berlinghieri F, Gentile A, Persichetti MF, Arculeo M, Alduina R (2019) New insights into the gut microbiome in loggerhead sea turtles Caretta caretta stranded on the Mediterranean coast. PLoS ONE 14:e0220329–e0220329. https://doi.org/10.1371/journal.pone.0220329
Biagi E, D’Amico F, Soverini M, Angelini V, Barone M, Turroni S, Rampelli S, Pari S, Brigidi P, Candela M (2019) Faecal bacterial communities from Mediterranean loggerhead sea turtles (Caretta caretta). Environ Microbiol Rep 11:361–371. https://doi.org/10.1111/1758-2229.12683
Filek K, Trotta A, Gračan R, Di Bello A, Corrente M, Bosak S (2021) Characterization of oral and cloacal microbial communities of wild and rehabilitated loggerhead sea turtles (Caretta caretta). Anim Microbiome 3:59. https://doi.org/10.1186/s42523-021-00120-5
Biagi E, Musella M, Palladino G, Angelini V, Pari S, Roncari C, Scicchitano D, Rampelli S, Franzellitti S, Candela M (2021) Impact of plastic debris on the gut microbiota of Caretta caretta from Northwestern Adriatic Sea. Front Mar Sci 8:637030. https://doi.org/10.3389/fmars.2021.637030
Scheelings TF, Moore RJ, Van TTH, Klaassen M, Reina RD (2020) The gut bacterial microbiota of sea turtles differs between geographically distinct populations. Endanger Species Res 42:95–108. https://doi.org/10.3354/esr01042
Kanjer L, Filek K, Mucko M, Majewska R, Gračan R, Trotta A, Panagopoulou A, Corrente M, Di Bello A, Bosak S (2022) Surface microbiota of Mediterranean loggerhead sea turtles unraveled by 16S and 18S amplicon sequencing. Front Ecol Evol 10:907368. https://doi.org/10.3389/fevo.2022.907368
Blasi MF, Rotini A, Bacci T, Targusi M, Ferraro GB, Vecchioni L, Alduina R, Migliore L (2021) On Caretta caretta ’s shell: first spatial analysis of micro- and macro-epibionts on the Mediterranean loggerhead sea turtle carapace. Mar Biol Res 17:762–774. https://doi.org/10.1080/17451000.2021.2016840
Scheelings TF, Moore RJ, Van TTH, Klaassen M, Reina RD (2020) Microbial symbiosis and coevolution of an entire clade of ancient vertebrates: the gut microbiota of sea turtles and its relationship to their phylogenetic history. Anim Microbiome 2:17–17. https://doi.org/10.1186/s42523-020-00034-8
Guo Y, Chen H, Liu P, Wang F, Li L, Ye M, Zhao W, Chen J (2022) Microbial composition of carapace, feces, and water column in captive juvenile green sea turtles with carapacial ulcers. Front Vet Sci 9. https://doi.org/10.3389/fvets.2022.1039519
Trevelline BK, Fontaine SS, Hartup BK, Kohl KD (2019) Conservation biology needs a microbial renaissance: a call for the consideration of host-associated microbiota in wildlife management practices. Proc R Soc B Biol Sci 286:20182448. https://doi.org/10.1098/rspb.2018.2448
Bjorndal KA, Bolten AB, Martins HR (2000) Somatic growth model of juvenile loggerhead sea turtles Caretta caretta: duration of pelagic stage. Mar Ecol Prog Ser 202:265–272. https://doi.org/10.3354/meps202265
Pinou T, Domenech F, Majewska R, Pfaller JB, Zardus JD, Robinson NJ (2019) Standardizing Sea Turtle Epibiont Sampling: Outcomes of the Epibiont Workshop at the 37 th International Sea Turtle Symposium. Mar Turt Newsl. https://doi.org/10.13140/RG.2.2.31843.81440
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:1–11. https://doi.org/10.1093/nar/gks808
White TJ, Bruns T, Lee S, Taylor J, AMPLIFICATION AND DIRECT SEQUENCING OF FUNGAL RIBOSOMAL RNA GENES FOR PHYLOGENETICS. In (1990) PCR protocols. Elsevier, pp 315–322
Andrews S (2010) FastQC: a quality control tool for high throughput sequencing data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R (2011) UniFrac: an effective distance metric for microbial community comparison. ISME J 5:169–172. https://doi.org/10.1038/ismej.2010.133
Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ (2017) Microbiome datasets are compositional: and this is not optional. Front Microbiol 8. https://doi.org/10.3389/fmicb.2017.02224
Martino C, Morton JT, Marotz CA, Thompson LR, Tripathi A, Knight R, Zengler K (2019) A novel sparse compositional technique reveals microbial perturbations. mSystems 4:1–13. https://doi.org/10.1128/mSystems.00016-19
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H (2020) vegan: Community Ecology Package. R package version 2.6-4. 2022 https://cran.r-project.org/web/packages/vegan/index.html
Arbizu PM (2017) pairwiseAdonis: Pairwise Multilevel Comparison using Adonis. R package version 0.4 2020 https://github.com/pmartinezarbizu/pairwiseAdonis
Peddada S, Lin H (2023) Multi-group analysis of compositions of Microbiomes with Covariate adjustments and repeated measures. In Review https://doi.org/10.21203/rs.3.rs-2778207/v1
Lin H, Peddada SD (2020) Analysis of compositions of microbiomes with bias correction. Nat Commun 11:3514. https://doi.org/10.1038/s41467-020-17041-7
Bisanz JE (2018) qiime2R: Importing QIIME2 artifacts and associated data into R sessions. R package version 0.99.6 2018 https://github.com/jbisanz/qiime2R
Wickham H, Averick M, Bryan J, Chang W, McGowan LD, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen TL, Miller E, Bache SM, Müller K, Ooms J, Robinson D, Seidel DP, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H (2019) Welcome to the tidyverse. J Open Source Softw 4:1686. https://doi.org/10.21105/joss.01686
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer https://doi.org/10.1007/978-0-387-98141-3
Reese AT, Dunn RR (2018) Drivers of Microbiome Biodiversity: a review of General rules. Feces Ignorance mBio 9. https://doi.org/10.1128/mbio.01294-18
Mariani G, Bellucci F, Cocumelli C, Raso C, Hochscheid S, Roncari C, Nerone E, Recchi S, Di Giacinto F, Olivieri V, Pulsoni S, Matiddi M, Silvestri C, Ferri N, Renzo LD (2023) Dietary preferences of Loggerhead Sea Turtles (Caretta caretta) in two Mediterranean Feeding grounds: does Prey Selection Change with Habitat Use throughout their life cycle? Anim Open Access J MDPI 13:654. https://doi.org/10.3390/ani13040654
Casale P, Abbate G, Freggi D, Conte N, Oliverio M, Argano R (2008) Foraging ecology of loggerhead sea turtles Caretta caretta in the central Mediterranean Sea: evidence for a relaxed life history model. Mar Ecol Prog Ser 372:265–276. https://doi.org/10.3354/meps07702
Hao W, Gerdts G, Peplies J, Wichels A (2015) Bacterial communities associated with four ctenophore genera from the German bight (North Sea). FEMS Microbiol Ecol 91:1–11. https://doi.org/10.1093/femsec/fiu006
Mabrok M, Algammal AM, Sivaramasamy E, Hetta HF, Atwah B, Alghamdi S, Fawzy A, Avendaño-Herrera R, Rodkhum C (2023) Tenacibaculosis caused by Tenacibaculum maritimum: updated knowledge of this marine bacterial fish pathogen. Front Cell Infect Microbiol 12:1068000. https://doi.org/10.3389/fcimb.2022.1068000
Gilbert MJ, Duim B, Zomer AL, Wagenaar JA (2019) Living in Cold blood: Arcobacter, Campylobacter, and Helicobacter in Reptiles. Front Microbiol 10:450049. https://doi.org/10.3389/fmicb.2019.01086
Albuquerque L, Simões C, Nobre MF, Pino NM, Battista JR, Silva MT, Rainey FA, de Costa MS (2005) Truepera radiovictrix gen. nov., sp. nov., a new radiation resistant species and the proposal of Trueperaceae fam. Nov. FEMS Microbiol Lett 247:161–169. https://doi.org/10.1016/j.femsle.2005.05.002
Tian Z, Pu H, Cai D, Luo G, Zhao L, Li K, Zou J, Zhao X, Yu M, Wu Y, Yang T, Guo P, Hu X (2022) Characterization of the bacterial microbiota in different gut and oral compartments of splendid japalure (Japalura Sensu lato). BMC Vet Res 18:205. https://doi.org/10.1186/s12917-022-03300-w
Hattenrath-Lehmann TK, Jankowiak J, Koch F, Gobler CJ (2019) Prokaryotic and eukaryotic microbiomes associated with blooms of the ichthyotoxic dinoflagellate Cochlodinium (Margalefidinium) polykrikoides in New York, USA, estuaries. PLoS ONE 14:e0223067. https://doi.org/10.1371/journal.pone.0223067
Li S, Young T, Archer S, Lee K, Alfaro AC (2023) Gut microbiome resilience of green-lipped mussels, Perna Canaliculus, to starvation. Int Microbiol. https://doi.org/10.1007/s10123-023-00397-3
Kasana RC, Pandey CB (2018) Exiguobacterium: an overview of a versatile genus with potential in industry and agriculture. Crit Rev Biotechnol 38:141–156. https://doi.org/10.1080/07388551.2017.1312273
Liu F, Li Y, He W, Wang W, Zheng J, Zhang D (2021) Exiguobacterium algae sp. nov. and Exiguobacterium qingdaonense sp. nov., two novel moderately halotolerant bacteria isolated from the coastal algae. Antonie Van Leeuwenhoek 114:1399–1406. https://doi.org/10.1007/s10482-021-01594-8
Cong M, Jiang Q, Xu X, Huang L, Su Y, Yan Q (2017) The complete genome sequence of Exiguobacterium arabatum W-01 reveals potential probiotic functions. MicrobiologyOpen 6:e00496. https://doi.org/10.1002/mbo3.496
Keller AG, Apprill A, Lebaron P, Robbins J, Romano TA, Overton E, Rong Y, Yuan R, Pollara S, Whalen KE (2021) Characterizing the culturable surface microbiomes of diverse marine animals. FEMS Microbiol Ecol 97:fiab040. https://doi.org/10.1093/femsec/fiab040
Filek K, Lebbe L, Willems A, Chaerle P, Vyverman W, Žižek M, Bosak S (2022) More than just hitchhikers: a survey of bacterial communities associated with diatoms originating from sea turtles. FEMS Microbiol Ecol 98:fiac104. https://doi.org/10.1093/femsec/fiac104
Jones EBG, Pang K-L, Abdel-Wahab MA, Scholz B, Hyde KD, Boekhout T, Ebel R, Rateb ME, Henderson L, Sakayaroj J, Suetrong S, Dayarathne MC, Kumar V, Raghukumar S, Sridhar KR, Bahkali AHA, Gleason FH, Norphanphoun C (2019) An online resource for marine fungi. Fungal Divers 96:347–433. https://doi.org/10.1007/s13225-019-00426-5
Cafarchia C, Paradies R, Figueredo LA, Iatta R, Desantis S, Di Bello AVF, Zizzo N, van Diepeningen AD (2020) Fusarium spp. in Loggerhead Sea Turtles (Caretta caretta): from colonization to infection. Vet Pathol 57:139–146. https://doi.org/10.1177/0300985819880347
Geiser DM, Al-Hatmi AMS, Aoki T, Arie T, Balmas V, Barnes I, Bergstrom GC, Bhattacharyya MK, Blomquist CL, Bowden RL, Brankovics B, Brown DW, Burgess LW, Bushley K, Busman M, Cano-Lira JF, Carrillo JD, Chang H-X, Chen C-Y, Chen W, Chilvers M, Chulze S, Coleman JJ, Cuomo CA, de Beer ZW, de Hoog GS, Del Castillo-Múnera J, Del Ponte EM, Diéguez-Uribeondo J, Di Pietro A, Edel-Hermann V, Elmer WH, Epstein L, Eskalen A, Esposto MC, Everts KL, Fernández-Pavía SP, da Silva GF, Foroud NA, Fourie G, Frandsen RJN, Freeman S, Freitag M, Frenkel O, Fuller KK, Gagkaeva T, Gardiner DM, Glenn AE, Gold SE, Gordon TR, Gregory NF, Gryzenhout M, Guarro J, Gugino BK, Gutierrez S, Hammond-Kosack KE, Harris LJ, Homa M, Hong C-F, Hornok L, Huang J-W, Ilkit M, Jacobs A, Jacobs K, Jiang C, Jiménez-Gasco M, del Kang M, Kasson S, Kazan MT, Kennell K, Kim JC, Kistler H-S, Kuldau HC, Kulik GA, Kurzai T, Laraba O, Laurence I, Lee MH, Lee T, Lee Y-W, Leslie Y-H, Liew JF, Lofton ECY, Logrieco LW, López-Berges AFS, Luque M, Lysøe AG, Ma E, Marra L-J, Martin RE, May FN, McCormick SR, McGee SP, Meis C, Migheli JF, Mohamed Nor Q, Monod NMI, Moretti M, Mostert A, Mulè D, Munaut G, Munkvold F, Nicholson GP, Nucci P, O’Donnell M, Pasquali K, Pfenning M, Prigitano LH, Proctor A, Ranque RH, Rehner S, Rep SA, Rodríguez-Alvarado M, Rose G, Roth LJ, Ruiz-Roldán MG, Saleh C, Salleh AA, Sang B, Scandiani H, Scauflaire MM, Schmale J, Short DG, Šišić DPG, Smith A, Smyth JA, Son CW, Spahr H, Stajich E, Steenkamp JE, Steinberg E, Subramaniam C, Suga R, Summerell H, Susca BA, Swett A, Toomajian CL, Torres-Cruz C, Tortorano TJ, Urban AM, Vaillancourt M, Vallad LJ, van der Lee GE, Vanderpool TAJ, van Diepeningen D, Vaughan AD, Venter MM, Vermeulen E, Verweij M, Viljoen PE, Waalwijk A, Wallace C, Walther EC, Wang G, Ward J, Wickes TJ, Wiederhold BL, Wingfield NP, Wood MJ, Xu AKM, Yang J-R, Yli-Mattila X-B, Yun T, Zakaria S-H, Zhang L, Zhang H, Zhang N, Zhang SX (2021) Phylogenomic Analysis of a 55.1-kb 19-Gene Dataset Resolves a Monophyletic Fusarium that Includes the Fusarium solani Species Complex. Phytopathology® 11:1064–1079
Hoh DZ, Lin Y-F, Liu W-A, Sidique SNM, Tsai IJ (2020) Nest microbiota and pathogen abundance in sea turtle hatcheries. Fungal Ecol 47:100964. https://doi.org/10.1016/j.funeco.2020.100964
Lavrinienko A, Scholier T, Bates ST, Miller AN, Watts PC (2021) Defining gut mycobiota for wild animals: a need for caution in assigning authentic resident fungal taxa. Anim Microbiome 3:75. https://doi.org/10.1186/s42523-021-00134-z
Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW (2014) Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. https://doi.org/10.1186/s12915-014-0087-z
Grahn N, Olofsson M, Ellnebo-Svedlund K, Monstein H-J, Jonasson J (2003) Identification of mixed bacterial DNA contamination in broad-range PCR amplification of 16S rDNA V1 and V3 variable regions by pyrosequencing of cloned amplicons. FEMS Microbiol Lett 219:87–91. https://doi.org/10.1016/S0378-1097(02)01190-4
Acknowledgements
For the Croatian sample collection, we are thankful to Milena Mičić, Karin Gobić Medica, and the rest of the staff from the Marine Turtle Rescue Center (Aquarium Pula). For the Italian collection of the samples and recruitment of turtles, we are thankful to Pasquale Salvemini of the “Centro di recupero tartarughe marine WWF Molfetta”.
Funding
This work has been fully supported by the Croatian Science Foundation under the project number UIP-2017-05-5635. The work of doctoral student K. Filek has been fully supported by the “Young Researchers’ Career Development Project – Training of Doctoral Students” of the Croatian Science Foundation funded by the European Union from the European Social Fund.
Author information
Authors and Affiliations
Contributions
K.F. and S.B. concepted and designed the study; A.T., M.C., A.B. and S.B. collected the samples; M.Ž., L.K. and K.F. carried out the laboratory work; K.F. and B.V. conducted the bioinformatics, statistical analyses, and data visualization and interpretation; K.F. wrote the first draft of the manuscript; all authors revised the paper and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethical Approval
Sampling was performed in accordance with the 1975 Declaration of Helsinki, as revised in 2013 and the applicable national laws. The sampling at the Sea Turtle Clinic (Bari, Italy) was conducted with the permission of the Department of Veterinary Medicine Animal Ethic Committee (Authorization # 4/19), while sampling in Croatia was done in accordance with the authorization of the Marine Turtle Rescue Center by the Ministry of Environment and Energy of the Republic of Croatia.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The EMBL ENA Accession number for one of our datasets was wrongly listed. It should be listed as PRJEB68298 in Material and Methods and Data Availability Sections.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Filek, K., Vuković, B.B., Žižek, M. et al. Loggerhead Sea Turtles as Hosts of Diverse Bacterial and Fungal Communities. Microb Ecol 87, 79 (2024). https://doi.org/10.1007/s00248-024-02388-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00248-024-02388-x