Abstract
Hot spring biofilms are stable, highly complex microbial structures. They form at dynamic redox and light gradients and are composed of microorganisms adapted to the extreme temperatures and fluctuating geochemical conditions of geothermal environments. In Croatia, a large number of poorly investigated geothermal springs host biofilm communities. Here, we investigated the microbial community composition of biofilms collected over several seasons at 12 geothermal springs and wells. We found biofilm microbial communities to be temporally stable and highly dominated by Cyanobacteria in all but one high-temperature sampling site (Bizovac well). Of the physiochemical parameters recorded, temperature had the strongest influence on biofilm microbial community composition. Besides Cyanobacteria, the biofilms were mainly inhabited by Chloroflexota, Gammaproteobacteria, and Bacteroidota. In a series of incubations with Cyanobacteria-dominated biofilms from Tuhelj spring and Chloroflexota- and Pseudomonadota-dominated biofilms from Bizovac well, we stimulated either chemoorganotrophic or chemolithotrophic community members, to determine the fraction of microorganisms dependent on organic carbon (in situ predominantly produced via photosynthesis) versus energy derived from geochemical redox gradients (here simulated by addition of thiosulfate). We found surprisingly similar levels of activity in response to all substrates in these two distinct biofilm communities, and observed microbial community composition and hot spring geochemistry to be poor predictors of microbial activity in the study systems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Geothermal aquifers are permeable layers of fluid-bearing rock that store some of the heat from the Earth’s interior [1]. Their high permeability allows continuous circulation of heat and fluids, but also the formation of temperature, chemical, and redox gradients [2, 3]. In consequence, hot springs function as autonomous bioreactors that allow colonization by diverse, specially adapted microbial communities (e.g., [4, 5]). These communities are often dominated by thermophilic Archaea and Bacteria that can survive in a temperature range of 50–121°C [6,7,8,9]. Extreme temperatures and pH have been shown to contribute most to limiting microbial diversity in geothermal aquifers and hot springs (e.g., [10,11,12,13]), although other environmental factors, such as light quality [14] and spring geochemistry [15,16,17], have also been shown to affect microbial community composition. Adaptation to the extreme conditions in hot springs requires genomic plasticity and metabolic flexibility of microorganisms, making them good candidates for the discovery of bioactive molecules of industrial and biotechnological interest [8, 18]. A well-known example is the discovery of Thermus aquaticus by T. Brock in the hot springs of Yellowstone National Park [19] and its DNA polymerase implementation in the polymerase chain reaction [20], which rapidly increased the interest in the study of microorganisms from geothermal springs.
Thermal springs are composed of various microenvironments such as sediments, water columns, microbial mats, and biofilms [21]. Biofilms are defined as associations of microbial cells, either of a single or multiple species, that have formed a community in an aggregated manner, attaching themselves to each other or to a surface by secreting extracellular polymeric substances (EPS) [22,23,24]. There is extensive literature describing the development of biofilms and the different phases of their establishment [24,25,26,27,28,29,30]. Hot spring biofilms have also been studied for decades and interest in them has still not waned. They are accessible, relatively stable, high biomass systems that harbor an enormous biotechnological potential [8, 31, 32]. Their composition is usually simple enough to be evaluated by traditional methods such as microscopy [33], but as they form on redox interfaces with well-defined environmental gradients, they can host a wide variety of metabolically diverse microorganisms (e.g., [34,35,36]). Although a large portion of biofilm-dwelling microorganisms can be isolated into pure culture, activities in pure cultures do not necessarily reflect in situ activities and do not provide insight into interactions within the biofilm community [37]. In summation, despite the comparatively low species richness, it is not trivial to understand the drivers behind the observed community structure and the biogeochemical cycling mechanisms due to the complex microbial interaction networks that form in hot spring biofilms [38].
The body of literature on the microbial community composition and the genomic landscape of hot spring biofilms continues to grow. However, despite the diverse suit of available methods to investigate the ecology and ecophysiology of biofilm microbial communities, such data are scarce for hot spring biofilms. In microbial ecology, incorporation of radio or stable isotope-labeled, or analogous substrates into microbial biomass is often used to trace metabolic activity [39]. Such substrate tracing approaches have been used to determine bulk activities of mixed biofilm communities (e.g., [21]) and to link specific metabolic activities to certain groups of hot spring biofilm organisms (e.g., via lipid biomarkers, [35, 40]). The same principle can also be used to link the utilization of a substrate to an individual cell via so-called single cell approaches [41,42,43]. Microbial cells that form hot spring (and other) biofilms are enclosed in an EPS matrix that restricts the diffusion of large molecules, and hinders the separation of cells and matrix components often necessary for downstream analysis, making the application of some physiology-targeted single-cell methods and analyses challenging [44]. Nevertheless, there is a plethora of studies investigating biofilm communities at single cell resolution (e.g., [45,46,47]). It is thus surprising that single-cell ecophysiological methods have almost never been applied to geothermal biofilms: only a few microautoradiography (MAR) experiments conducted on photosynthetic hot spring biofilms and mats from different pools in Yellowstone have been published to date [48,49,50,51].
MAR visualizes the incorporation of radiolabeled isotopes into microbial cells and can be combined with fluorescence in situ hybridization (FISH) to identify active microorganisms, but sample processing is time consuming and requires specialized laboratory equipment and radioactive waste management [39, 42, 52]. Nanoscale secondary ion mass spectrometry (nanoSIMS) enables the visualization of stable isotope incorporation, circumventing the waste and safety issues related to working with radioisotopes, but low throughput and the need for sophisticated instrumentation limit its application in practice [39, 42, 52]. In addition, nanoSIMS only examines the surface of a sample, and although tomography is possible [53, 54], it is not practical for biofilms. Notably, both MAR and nanoSIMS are destructive methods that prevent downstream analyses, such as genome sequencing or cultivation [42]. Bioorthogonal non-canonical amino acid tagging (BONCAT), which can be used to visualize a large diversity of translationally active cells and can reveal under which conditions cells and cell populations are active [55, 56], has fewer drawbacks than other currently available single-cell methods. BONCAT is based on the incorporation of azide- or alkyne-group containing l-methionine analogs, whereupon newly synthesized proteins can be detected using azide-alkyne click chemistry [57]. BONCAT requires a relatively simple infrastructure available to most laboratories and is a rapid and nondestructive method compatible with many other approaches such as (meta)genomics, (meta)proteomics, nanoSIMS, fluorescence activated cell sorting (FACS), and FISH [39, 42, 58,59,60]. The combination of BONCAT with FISH allows to directly link the identity and activity of microorganisms in environmental samples by fluorescence microscopy [45]. Albeit being a relatively novel method, it has already been successfully applied on various environmental sample types, including biofilms [45, 52, 56, 59, 61].
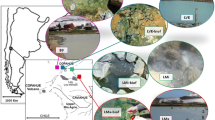
In this study, we applied a combination of catalyzed reported deposition (CARD) FISH and BONCAT to investigate the identity and activity of microorganisms inhabiting two contrasting hot spring biofilms, supplied with substrates stimulating either organotrophic or lithotrophic microbial activity. First, we applied 16S rRNA gene amplicon sequencing to screen the microbial community composition in biofilms collected from geothermally active sites in northern and northeastern Croatia (Fig. 1). This region, characterized by a high average geothermal gradient (49°C/km), surface heat flow (76 mW/m2), and shallow Mohorovičić discontinuity, hosts a large number of geothermal sites [62, 63], many of which contain biofilms that have not been previously studied. In a recent study [63], we characterized the hydrochemistry of the geothermal springs and investigated the microbial community composition of geothermal waters. However, the environmental conditions of geothermal waters differ significantly from the microenvironment in biofilms, where nutrients are entrapped and microorganisms form metabolically and functionally integrated consortia [64, 65]. In consequence, distinct microbial communities can be expected to occur in geothermal biofilms. To gain insight into the diversity and metabolic activity of biofilm microorganisms in hot springs, we initially characterized the microbial community of biofilms collected from 12 geothermal sources over three sampling seasons. Subsequently, two distinctly colonized biofilms from geothermal sources with different geochemistry were supplied with substrates that stimulated either chemoorganotrophic or thiosulfate-dependent chemolithotrophic microbial activity. Although we were able to confirm the contribution of select microbial community members to these metabolic processes, the results of the incubation experiments did not fully align with hypothesized activities derived from microbial community composition and thermal water chemistry data. Our findings underline how difficult it is to correctly infer microbial functions from taxonomic data, even in relatively simple systems such as hot spring biofilms.
Materials and methods
Sampling and Physicochemical Measurements
Up to three samplings of biofilms from 12 geothermal sources located at 10 geothermally active sites in Croatia (Fig. 1a; Table S1) were conducted in autumn 2019, spring 2020, and autumn 2020, while additional biofilm samples from two sites (the Bizovac well and the Tuhelj spring) were collected in spring 2021 (Table S1). Depending on the presence and localization of biofilms, samples were collected in a depth range from the air–water interface to up to 0.5 m water depth, using a sterile spatula and tweezers. The biofilm samples were stored in sterile 50-mL tubes and submerged in the corresponding geothermal water. If multiple different colored biofilms were present at one site (e.g., Fig. 1b), they were sampled separately. Likewise, multilayered biofilms were separated according to the layer color and further processed separately. After sampling, electrical conductivity (EC), dissolved oxygen (O2) concentration, pH, and water temperature in geothermal spring waters were measured in situ using a WTW multiparameter probe (WTW, Germany) (Table S1). Sampling of biofilms and in situ measurements of psychochemical parameters in geothermal spring waters were conducted as close as possible to the spring source, and the measured parameters reflect the geochemistry experienced by the sampled biofilms. The concentrations of hydrogen sulfide (H2S) in spring waters directly adjacent to or above the biofilms were determined using Reagent sulfide 1 and Reagent sulfide 2 according to the USEPA Methylene Blue Method with a HACH DR3900 spectrometer, as specified in DR manual 1 (https://www.hach.com/asset-get.download-en.jsa?id=7639983731) (Table S1). If necessary, samples were diluted for H2S concentration measurements. For further chemical analysis in the laboratory, water samples were collected in 50-mL tubes and 500-mL plastic bottles. All samples were brought to the laboratory within a few hours after collection. Chemical analyses were performed at the Croatian Geological Institute (see [63] and Supplementary Material for further details). The concentrations of cations (sodium, potassium, calcium, ammonium, and magnesium) and anions (chloride, nitrate, and sulfate), alkalinity, SiO2 concentrations, and dissolved organic (DOC) and inorganic (DIC) carbon concentration were determined the same evening after returning from the field site (Table S1). The results of these chemical analyses were in parts already published previously in [63], where a detailed description of sampled geothermal sites can also be found.
Biofilm DNA Extraction, 16S rRNA Gene Amplification, Sequencing, and Data Processing
After excess water was removed from the collected biofilm samples by decanting and pipetting, 0.5 g of each biofilm sample was weighed into beat beating tubes for DNA extraction. According to the manufacturer’s guidelines, total genomic DNA was extracted using the DNeasy PowerSoil Kit (Qiagen GmbH Hilden, Germany). The hypervariable V4 region of the 16S rRNA gene was performed using primer pairs 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) [66] and 806R (5′-GGACTACNVGGGTWTCTAAT-3′) [67] at the Joint Microbiome Facility of the Medical University of Vienna and the University of Vienna (project IDs JMF-2007-4, JMF-2103-13, JMF-2110-16). Samples were amplified, barcoded, purified, normalized, and prepared for sequencing as previously described [68] and sequenced in pair-end mode (v3 chemistry, 2 × 300 bp) on an Illumina MiSeq instrument. Amplicon pools were extracted from raw sequencing data using default FASTQ workflow parameters (BaseSpace; Illumina), filtered for PhiX contamination using BBDuk (BBtools) [69, 70], and demultiplexed using the Python package demultiplex (Laros JFJ, github.com/jfjlaros/demultiplex), allowing for one mismatch per barcode and two mismatches for linker and primer sequences. FASTQ reads were trimmed at 220 and 150 nt, respectively, with an error of 2 allowed for forward and reverse reads each. Amplicon sequence variants (ASVs) were inferred in pooled mode in the DADA2 R package version 1.20.0 (R 4.1.1, [70, 71]) using standard settings, and classified against the SILVA reference database (SILVA release 138) using the SINA classifier, version 1.6.1 [72]. Prior to downstream analysis, ASVs classified as eukaryotes, mitochondria, or chloroplasts, as well as unclassified ASVs were removed from the dataset, as were singletons and doubletons. After filtering, samples with less than 3000 sequence reads were excluded from the dataset. The final ASV table contained 4201 ASVs and 59 samples with read counts greater than 3000. All statistical analyses were performed in the R environment (v. 4.2.0) using Bioconductor v3.15 packages TreeSummarizedExperiment [73], phyloseq [74], mia [75], and microViz [76] and visualized using the package ggplot2 [77]. The similarity of prokaryotic community between biofilm samples as a function of their environmental parameters was tested using non-metric multidimensional scaling (NMDS) based on Bray–Curtis dissimilarity distance. Permutational multivariate analysis of variance (PERMANOVA) was used to test the extent to which microbial communities were affected by sampling seasons, locations, selected environmental parameters, and biofilm color.
Data Availability
16S rRNA gene amplicon sequencing data have been deposited in SRA (the Sequence Read Archive) under the BioProject accession number PRJNA889237.
Incubation Experiments for Bizovac Well and Tuhelj Spring Biofilms
Fresh biofilm samples were collected in spring 2021 from the Bizovac well, an atypical biofilm not dominated by Cyanobacteria from a moderately sulfidic, high-temperature geothermal source, and from the Tuhelj spring, representing a Cyanobacteria-dominated biofilm from a low-sulfide system (Fig. 3; Table S1). We characterized the biofilm microbial community of these freshly collected samples by 16S rRNA gene amplicon sequencing and confirmed that it resembles the composition of the previously analyzed biofilm samples collected in 2019 and 2020 from the respective source environment (Fig. 3a; Fig. S1). To assess the activity of microorganisms in hot spring biofilms sustained by different reducing equivalent sources, we designed a set of incubation experiments that stimulated either aerobic chemoorganotrophic or chemolithotrophic microbial activity. Since the redox gradients in the here investigated hot springs, including Bizovac well and Tuhelj spring, were partially characterized by the presence of sulfide-oxygen transitions zones ([63], Table S1), we provided thiosulfate as reducing equivalent source for chemolithotrophic microorganisms in our experiments. Unlike sulfide, thiosulfate is stable under oxic conditions and non-toxic at elevated concentrations, and the majority of sulfide oxidizers can also utilize thiosulfate [78]. Glucose or acetate amendments were used as organic substrates and reducing equivalents, as previous studies have shown that both glucose and acetate are released by Cyanobacteria as products of glycogen degradation under dark and anaerobic conditions [79, 80]. The combination of BONCAT and CARD-FISH was performed as described below (Fig. 2). For each incubation setup (Fig. 2), biofilm material (0.5 g wet weight) was suspended in 10 mL sterile filtered (0.2 μm pore size) corresponding geothermal spring water in glass vials and incubated for 48 h at source temperature. To monitor activity, l-homopropargylglycine (HPG) was added to the sample at a final concentration of 50 μmol L−1 alongside substrates (sodium acetate, glucose, and sodium thiosulfate) at a final concentration of 1 mmol L−1. Acetate and glucose solutions were prepared by dissolving the appropriate weight of substrate in autoclaved MQ water, while the thiosulfate media stock was prepared according to [81]. To assess native activity, a set of incubations was also performed without substrate amendment, with HPG addition only. All incubations were performed in the dark to minimize the effect of additional photosynthetically produced substrates on the experiment. After 24 and 48 h, samples were removed from the incubator and 2 mL of 0.5% Tween 20 (Promega) was added [82]. Samples were then macerated, vortexed at maximum speed for 5 min, sonicated at standard instrument settings for 1 min (Microson XL 2000) [83], and centrifuged at 500 × g for 5 min to separate the particles from the cell fraction [82]. The supernatant was sequentially filtered through 10 μm and 3 μm pore size polycarbonate filters to remove filamentous Cyanobacteria. The final filtrate was fixed with paraformaldehyde at a final concentration of 3% for 1 h, at room temperature, in the dark [83]. Samples were then filtered onto a 0.2 μm pore size filter, examined under the microscope using 4′,6-diamidino-2-phenylindole (DAPI) stain (10 μg μL−1), and the filters were stored at −20°C until the click reaction was performed.
A schematic overview of the experimental workflow applied for hot spring biofilm characterization in this study: (1) characterization of the biofilm microbial community composition; (2) incubation with the l-methionine analogue HPG under different substrate amendment in the dark, for 24 h and 48 h and BONCAT visualization of active cells; (3) CARD-FISH hybridization of target populations, sample imaging and image analysis
BONCAT was performed according to [57]. Briefly, filter pieces were placed sequentially in 50, 80, and 96% EtOH for 3 min to dehydrate and permeabilize the cells. For the Cu(I)-catalyzed click reaction, a dye premix was prepared containing 1.25 μL of a 20 mmol L−1 CuSO4 solution, 1 μL of a 2.5 mmol L−1 Cy5-alkyne dye, and 2.5 μL of a 50 mmol L−1 tris-hydroxypropyltriazolylmethylamine chelating agent. While allowing the dye-premix to react, 12.5 μL of each 100 mmol L−1 sodium ascorbate and 100 mmol L−1 aminoguanidine hydrochloride were added to 221 μL phosphate buffered saline (PBS). After mixing the dye-premix with the rest of the buffer, the filters were added directly to the click mixture and incubated at room temperature in the dark for 1 h. The filters were then washed three times in PBS for 3 min each, dehydrated in 50% EtOH for 3 min, and stored at −20°C until the CARD-FISH procedure.
CARD-FISH was performed according to [84]. Briefly, after the filters thawed, they were dehydrated in a series of EtOH and embedded in 0.1% low melting point agarose. Permeabilization was performed with a 10 mg mL−1 lysozyme solution for 1 h and 60 U mL−1 achromopeptidase solution for 30 min, both at 37°C, followed by incubation in 0.1 mol L−1 HCl for 1 min at room temperature. Inactivation of endogenous peroxidases was achieved by 3% H2O2 treatment. Subsequently, 1.5 μL of a 50 ng μL−1 HRP-probe solution was added to 400 μL of hybridization buffer of corresponding stringency (Table S2). Filters were added directly to the solution and hybridized overnight at 46°C. To improve detection of microorganisms, the tyramide signal was amplified by catalyzed reporter deposition (CARD) [85]. Before tyramide signal amplification, washing was performed in washing buffer at 48°C for 10 min in a water bath and in Triton-X–PBS for 5 min. Filters were placed in a mixture of 1 mL of amplification buffer and 2 μL of tyramide solution (either Cy3 or Atto-488), incubated at 46°C for 20 min, and washed in Triton-X–PBS, MQ water, and absolute EtOH. After drying, filters were placed on the slide and embedded in Citifluor Vectashield DAPI (10 μg μL−1) solution. Oligonucleotide probes for CARD-FISH were selected based on 16S rRNA gene amplicon analysis of biofilms from previous seasons (Table S2). The NONEUB probe was used as a negative control for all hybridizations.
Image Processing and Analysis
Images of the samples were acquired using an epifluorescence microscope (model DMi8; Leica, Germany) equipped with the Thunder Imaging System at a resolution of 2048 × 2048 pixels. For each sample, Z-stacks of 4–10 fields of view were acquired, in each channel individually as well as their overlay. The final images represent the maximum projection of all stacks. After inspection of the samples on the microscope, biovolume analysis was performed using daime software (v. 2_2_3) [86]. Images were processed with 2D filter histogram stretching to remove any background noise before automatic segmentation in ISODATA or Custom mode. After examining the results of various segmentation methods for each sample, the biovolume fractions of CARD-FISH positive cells/cell aggregates were determined against BONCAT positive ones, and vice versa. Results acquired through the daime software can be found in Table S2.
Results
Geothermal Spring Characteristic and Seasonal Biofilm Sampling
The here sampled geothermal springs and wells (n = 12) differed significantly in terms of their geochemistry (Table S1, [63]). In general, samples from the well Topusko had the highest temperature (TEB, 64.8°C) while samples from the D. Breznica well (DB, 12.9°C) had the lowest temperature. The pH ranged from 6.3 to 8.3, DIC from 52 to 390 mg L−1, and DOC from 0.05 to 35.5 mg L−1 (Table S1). The highest concentration of H2S was observed in spring 2020 in the DB well (18.8 mg L−1), while concentrations of SO42− ranged from 9.3 to 196.9 mg L−1 (Table S1). Samples from the Zelina and Bizovac well had the highest NH4+ concentration (9.03, 8.82 mg L−1). A more detailed description and characterization of sampling sites according to their environmental parameters can be found in [63].
Based on 16S rRNA gene amplicon sequencing, the microorganisms inhabiting 59 biofilm (sub)samples collected over three seasons were clustered into 4201 ASVs. NMDS analysis showed that the microbial community of the biofilms mainly clustered by spring or well of origin (Fig. 3a). This result was verified by PERMANOVA, confirming source spring as a significant factor affecting biofilm community composition (R2 = 0.42, P < 0.001). Among the measured environmental parameters, the community structure was best explained by temperature (R2 = 0.8, P < 0.001) and electric conductivity (which correlated to the concentrations of sodium, potassium, and chlorine ions) (R2 = 0.5, P < 0.001) (Table S3). The season of sampling (R2 = 0.06, P = 0.08) and the color of the biofilm (sub)samples (R2 = 0.08, P = 0.89) did not significantly correlate with observed community structure at the ASV level.
(a) NMDS clustering of hot spring biofilm microbial communities. Samples are colored by sampling location, while shapes represent the sampling season. (b) Microbial community composition clustered at phylum level. Phyla were cumulative relative abundance of ASVs exceeded 5% in at least one biofilm (sub)sample are displayed, while the residual community is clustered as “other.” Samples are grouped based on the color of biofilm (sub)samples. Sampling locations are indicated by the sample site abbreviations, while shapes represent the sampling season
Overall, the biofilm communities were highly diverse, with ASVs affiliating with 52 bacterial and 10 archaeal phyla. While geothermal waters at the same sampling site were dominated by ASVs affiliating to Crenarchaeota, Nanoarchaeota, Campylobacterota, and various Pseudomonadota lineages [63], almost all here analyzed biofilm samples were dominated by Cyanobacteria. Pseudomonadota, Chloroflexota, and Bacteroidota also displayed high relative abundances in almost all biofilm samples (Fig. 3b). The relative abundance of Archaea was comparatively low in all samples (7 ± 3% on average). Unlike most samples, biofilms collected from the Bizovac well were not dominated by Cyanobacteria (Fig. 3b). Instead, Pseudomonadota, Chloroflexota, Deinococcota, and Bacteroidota accounted for large fractions of the community composition in biofilm samples from this site. At the family and genus level, prokaryotic community composition was highly site-specific (Fig. S1): Leptolyngbyaceae and Nostocaceae were the relatively most abundant cyanobacterial families in all biofilms, except for those collected at the highly sulfidic low temperature well DB, where Trychonema-related ASV (family Phormidiaceae) dominated the cyanobacterial community. In samples from Bizovac well, Cyanobacteria were not only relatively less abundant overall, but also distinct from cyanobacterial communities of the remaining biofilms, with ASVs predominantly affiliating with unicellular, colony-forming members of the genus Gloeocapsa (family Chroococcidiopsaceae; Fig. S1). In general, Bizovac well samples displayed a relatively high diversity, with few highly abundant individual genera and rather distinct community composition compared to samples from other geothermal sites (Fig. 3; Fig. S1). Among thiotrophic community members, unicellular gammaproteobacterial Halothiobacillaceae (genus Thiofaba) dominated biofilms from the Bizovac well during all three sampling seasons, whereas filamentous gammaproteobacterial population (family Thiotrichaceae, genus Thiothrix; Fig. S1) and ASVs related to the genus Sulfurovum (Campylobacterota; Fig. S1) were detected in the samples from all other sites.
The initial community of freshly collected (spring 2021) biofilms from the Bizovac well used for the activity experiments was again dominated by Bacteroidota (28.1%), Pseudomonadota (27.3%), and Chloroflexota (11.1%), while biofilms from the Tuhelj spring were again dominated by Cyanobacteria (41.5%), but Pseudomonadota (13.8%) and Chloroflexota (14.7%) were also relatively abundant (Fig. 3b; Fig. S1).
Chemoorganotrophic versus Chemolithotrophic Microbial Activity of Biofilms from Bizovac Well and Tuhelj Spring
As expected, the active fraction of the bacterial biofilm community (defined as displaying a CARD-FISH signal with the probe mixture EUBI-III targeting the majority of bacteria, and a positive BONCAT signal) was lowest when the biofilm samples were incubated without substrate amendment (i.e., HPG control). The active fraction of cells in the incubations without substrate amendment was very similar in both biofilm samples: after 24 h of incubation, 33% and 35% of EUBI-III positive cells displayed a BONCAT signal in Bizovac and Tuhelj biofilm incubations, respectively (Fig. 4). Furthermore, the fraction of active cells in substrate amendment free controls did not change significantly after 48 h of incubation in either biofilm (Fig. 4; Table S4), indicating that native energy and substrate sources have been nearly depleted, or that all microorganisms capable of utilizing these under the applied experimental conditions became active within 48 h. The 48-h incubation time point was thus selected to further inspect the effects of different substrate amendments on the fraction and identity of the active biofilm populations.
Biovolume of the active fraction (i.e., displaying BONCAT signal) of cells displaying a positive CARD-FISH single with probes targeting specifically the Chloroflexota (CFX1223), the Gammaproteobacteria (Gam42a), or all bacteria (EUBI-III) after 24 h without substrate amendment (HPG only), and 48 h without substrate amendment and in incubations amended with glucose, acetate or thiosulfate
Glucose, acetate, and thiosulfate amendments all resulted in a significant increase of the active bacterial fraction (as defined by positive signal with the EUBI-III probe, and a positive BONCAT signal) after 48 h of incubation (Fig. 4). The glucose amendment resulted in the highest total active cell biovolume in the Bizovac biofilms (58%) and the thiosulfate amendment had the greatest effect on the total active cell fraction in the Tuhelj biofilms (62.5%), while the activity stimulated by acetate and glucose amendments was much lower (Fig. 4; Table S4). Moreover, the activity stimulated by acetate and glucose amendment in Tuhelj biofilms was lower than the activity stimulated by the same substrates in Bizovac biofilms (Fig. 4; Table S4). Because Gammaproteobacteria- and Chloroflexota-related ASVs were the most abundant non-cyanobacterial ASVs in both biofilms (Fig. S2), the contributions of these taxa were selected for a more in-depth analysis of the observed responses to substrate amendment by BONCAT–CARD-FISH.
Gammaproteobacteria accounted for 22.8% of the initial population in Bizovac biofilms, including high relative abundances of the families Rhodocyclaceae (10.6%), Hallothiobacillaceae (4.9%), and Hydrogenophilaceae (1.8%), known to harbor thiosulfate oxidizers (Fig. S2). In the Tuhelj biofilms, Gammaproteobacteria-related ASVs accounted for 11.2% of the total microbial population, with most ASVs belonging to the families Rhodocyclaceae (3.6%), Chromatimaceae (1.1%), and Beggiatoaceae (1.0%) (Fig. S2). The fraction of active Gammaproteobacteria cells (defined as displaying a CARD-FISH signal with the Gam42a probe, and a positive BONCAT signal) also remained constant when comparing 24- and 48-h incubations without substrate amendment (Fig. 4). Under these conditions, ~25% of the gammaproteobacterial cells were active in the Bizovac biofilms, whereas only ~10% of the gammaproteobacterial population was active in Tuhelj samples (Table S4; Fig. 4). Glucose and thiosulfate amendments resulted in very similar responses of Gammaproteobacteria in Bizovac biofilms, with 56% and 61% of translationally active cells after 48 h of incubation, respectively (Fig. 4; Table S4). The acetate amendment had a negative effect on the activity of gammaproteobacterial cells in the Bizovac biofilms, and a BONCAT signal was detected in only 12.5% of the Gam42-positive cells after 48 h of incubation (Fig. 4; Table S4). In the Tuhelj biofilms, thiosulfate amendment resulted in a large increase of the active fraction of gammaproteobacterial cells, with 48% of translationally active Gam42a-positive cells after 48 h of incubation (Fig. 4; Table S4). The glucose amendment also resulted in increased activity of Gammaproteobacteria in the Tuhelj biofilms, with 36% of cells displaying activity after 48 h (Fig. 4; Table S4). As observed previously for the Bizovac biofilms, acetate amendment also led to a suppression of gammaproteobacterial activity in the Tuhelj biofilms. After 48 h of incubation with acetate, only 3.6% of gammaproteobacterial cells were translationally active (Fig. 4; Table S4).
In the Bizovac biofilms, Chloroflexota had a lower relative abundance than Gammaproteobacteria. Almost all ASVs affiliated to Chloroflexota (11.1%) belonged to the SBR1031 clade (4.4%) or the family Anaerolineaceae (6.1%) (Fig S2). In Tuhelj spring biofilm sample, Chloroflexota-related ASVs displayed a higher relative abundance then Gammaprotobacteria and accounted for the majority of the non-cyanobacterial microbial community (14.7%). Most ASVs in these samples were affiliated to the families Chloroflexaceae (8.9%) and Anaerolineaceae (2.9%) (Fig. S2). Less than 2% of Chloroflexota cells (defined as displaying a CARD-FISH signal with the CFX1223 probe, and a positive BONCAT signal) were active after 24 h, while 9% of cells were active after 48 h of incubation without substrate amendment in Bizovac biofilms (Table S4; Fig.4). In the Tuhelj biofilms, the activity of Chloroflexota cells in treatments without substrate amendment was much higher than in Bizovac biofilms, with 19% and 15% of active CFX1223-positive cells after 24 h and 48 h of incubation, respectively (Fig. 4; Table S4). Like the activity in the control treatment (no substrate amendment), the response of Chloroflexota populations in the Bizovac biofilms to glucose and thiosulfate was lower than the response of the gammaproteobacterial population. After 48 h of incubation in these substrates, 30% and 29% of the population were determined active, respectively (Fig. 4; Table S4). However, unlike Gammaproteobacteria, the Chloroflexota population in the Bizovac biofilms did respond with increased activity to acetate amendment, with 21% of active Chloroflexota cells after 48 h of incubation (Fig. 4; Table S4). In contrast, incubation with acetate in the Tuhelj biofilms resulted in lower activity of Chloroflexota compared with the control treatments. After 48 h of incubation with acetate amendment, only 9% of Chloroflexota cells were active (Fig. 4; Table S4). On the other hand, glucose amendments resulted in the highest increase in activity and percentage of active Chloroflexota cells in Tuhelj biofilms (47% after 48 h of incubation). Lastly, 37% of the Cholorflexota populations in the Tuhelj biofilms were active after 48 h of incubation in thiosulfate amended setups (Fig. 4; Table S4).
Discussion
The microbial composition of hot spring biofilms has often been described to be mainly influenced by the elevated temperature in these environments (e.g., [7, 24, 28, 87]). However, a study of 925 hot springs in New Zealand showed that temperature has a significant effect on microbial beta diversity only at values above 80°C [88]. Although geothermal spring temperatures were lower in this study (max. 65°C), temperature, together with the sample origin, were the main determinants of biofilm microbial structure (Table S3). In contrast, no significant effect of sampling season or biofilm color on microbial community composition was observed. The most abundant microbial groups found in the sampled biofilms were consistent with the literature [89,90,91,92,93,94,95,96], with Cyanobacteria dominating the biofilms of all but one hot spring (Bizovac well), and Chloroflexota, Pseudomonadota, and Bacteroidota displaying a high relative abundance in all biofilms. The Leptolyngbya-related cyanobacteria, which dominated most of the biofilms we analyzed, also dominated the upper layer of alkaline microbial mats from the Garga hot spring [97], while the genus Tychonema, which we found in biofilms from the low-temperature well DB, was recently found abundant in biofilms from hydrothermal systems in Italy with temperatures moderately above ambient [98]. As expected, the microbial communities in biofilms were clearly distinct from the geothermal waters communities in the same springs. For example, Archaea were not abundant in biofilm sample (Fig. 3), while diverse crenarchaeal, micrarchaeal, and nanoarchaeal lineages dominated many of the water samples collected form the same springs [63]. The highest abundance of Creanarchaeota was found in the biofilm from the Topusko well (TEB) the sampling site with the highest temperature (64.8°C) in this dataset. Comparison of mats from a geothermal spring in Romania at different temperature ranges showed that the relative abundance of Archaea was low at 32°C (<0.5%), but increased significantly at 65°C (36%) [91], which is in line with our observations. Similarly, Alcaman et al. reported a temperature value of 58°C as the turnover point for phototrophs, with Cyanobacteria dominating the lower temperature range and Chloroflexota being more abundant in the 58–66°C range [93]. Three of our high-temperature sampling sites (Topusko well (TEB), Varazdin well (V), and Bizovac well (B)) displayed temperatures around this tipping point (Table S1), but only biofilm samples from Bizovac well were not dominated by Cyanobacteria and had high relative abundances of Chloroflexota (genera Candidatus Chlorotrix and Chloroflexus; Fig. S1). All other high- and moderate-temperature sampling sites were in fact dominated by Cyanobacteria (Fig. 3b), confirming that temperature is not the only driver for the previously observed community switch among primary producers. Also, thiotrophs detected in water samples from the same sites (e.g., Sulfuricurvum and Sulfurimonas among the Camplyobacterota, and Thiobacillus among the Gammaproteobacteria, [63]) differed from the dominant genera in our biofilm samples (Sulfurovum, Thiofaba, Thiotrix, Hydrogenophillus), all previously reported to occur in hot spring biofilms from other sites with similar temperatures (e.g., [99]).
Based on the differences in microbial community composition and spring geochemistry observed during seasonal sampling, fresh biofilms were collected from the Bizovac well and the Tuhelj spring, and subjected to a series of incubation experiments to investigate the organo- and lihtotrophic activity of the non-photosynthetic community members. The majority of hot spring biofilms we analyzed, including those from Tuhelj spring, were dominated by Cyanobacteria, the dominant photosynthetic primary producers in these systems. Thus, Cyanobacteria are likely a major source of organic carbon substrates, sustaining growth and activity of chemoorganotrophic microorganisms residing in the hot spring biofilms. However, as the hot spring biofilms grow at a redox interface, a fraction of the microbial community might also be adapted to derive energy from reduced inorganic substrates via chemolithotrophic metabolisms, as evidenced, for example, by the presence of various and diverse thiotrophs in most biofilm samples (Fig. S1). Therefore, thiosulfate was selected as substrate to test chemolithotrophic activity related to the geochemical gradients the biofilms reside on. Glucose and acetate were selected as carbon substrates to simulate activity of chemoorganotrophs, in situ supplied with organic compounds via photosynthetic primary production. In biofilms recovered from both sources, the thiotrophs putatively able to oxidize the thiosulfate provided as substrate were predominantly affiliated with various gammaproteobacterial lineages.
The Bizovac biofilm included relatively more ASVs affiliated with putative thiotrophs and Gammaproteobacteria in general compared to Cyanobacteria-dominated biofilms from the Tuhelj spring (Fig. S2). ASVs related to Chromatiaceae and Hallothiobacillaceae were identified as putative thiotrophs in the Bizovac sample, while Chromatiaceae and Beggiatoaceae ASVs were found in the biofilms from the Tuhelj spring (Fig. S2; [100]). Other abundant gammaproteobacterial families in both samples were Rhodocyclaceae and Hydrogenophilaceae (Fig. S2). The family Rhodocyclaceae is metabolically diverse and consists of photoautotrophic, heterotrophic, and organotrophic members. This family has also been found abundantly in sulfidic hot springs in northern Baikal [101] and in microbial mats of hot springs in Eritrea [102]. The most abundant ASVs of the family Rhodocyclaceae detected in the Bizovac biofilm belonged to the genus Azoarcus (8%). Although primarily classified as denitrifying chemolithoheterotrophs, numerous studies reported Azoarcus to be core denitrifying desulfurizing bacteria in bioreactor experiments under mixotrophic conditions [103,104,105,106,107]. The only currently known sulfur-oxidizing denitrifying species, Azoarcus taiwanensis, was isolated from a hot spring biofilm in Taiwan [108]. Members of the family Hydrogenophilaceae are thermophilic heterotrophs and autotrophs that can use hydrogen as an electron donor [109]. A new species of this family was recently isolated from microbial mats of hot springs in Japan that can grow with thiosulfate and elemental sulfur under aerobic conditions and is unable to utilize hydrogen [110]. Almost all Hallothiobacilaceae ASVs in the Bizovac sample belonged to the genus Thiofaba (4.8%; Fig. S1). The type strain of this genus, Thiofaba teidiphila, was isolated from the biofilm of a hot spring in Japan and characterized as an obligate chemolithoautotrophic bacterium that utilizes thiosulfate, elemental sulfur, sulfide, and tetrathionate as electron donors [111]. Furthermore, a larger fraction of the microbial community in Bizovac biofilms was related to Desulfobacterota—a phylum harboring various microorganisms involved mainly in reductive, but also oxidative sulfur cycling [100]. Also, appreciable amounts of sulfide were detectable in situ in the water samples from Bizovac well, but not in waters of the Tuhelj spring (Table S1), indicating sulfide oxidation, but also S-disproportionation [112]. Hence, we hypothesized that either a larger fraction of the microbial community in the Bizovac biofilms would be capable of chemolithotrophic activity utilizing thiosulfate as source of reducing equivalents, or that the observed changes in the activity of the non-thiotrophic fraction, despite the relatively short incubation time, were the results of cross-feeding interactions in which the activity of the putative thiotrophs led to the accumulation of hydrogen and small organic molecules. Contrary to our hypothesis, the overall increase in the total active cell fraction and, in particular, the active gammaproteobacterial cell fraction was more pronounced in the Tuhelj biofilms in incubations amended with thiosulfate (Fig. 4), in which ASVs related to known thiotrophs were relatively less abundant (Fig. S2). Because the community in Bizovac biofilms was dominated by chemolithoheterotrophic Rhodocyclaceae, it is possible that the availability of suitable organic carbon sources limited the metabolic activity of thiotrophs in our experiments.
In both biofilm samples, the response to glucose amendments in the gammaproteobacterial cell fraction was in the same range of activity increase as in the thiosulfate amended incubations (Fig. 4). These results are in line with chemoorganoheterotrophy and mixotrophy generally expected for many of the community members detected here. The class Gammaproteobacteria contains the largest diversity of obligate and generalist hydrocarbonoclastic bacteria [113], many of which are known for their implication in acetate assimilation [114, 115]. However, contrary to our expectation, incubation with acetate resulted in decreased fractions of active gammaproteobacterial populations in both biofilms. Compared to glucose, acetate is a low-energy carbon source and has been shown to have an inhibitory effect on some microorganisms, reducing their growth rate [116]. However, as the overall response of the microbial community to acetate amendment was increased activity, the used acetate concentration does not appear to be generally inhibitory.
In both biofilms, the increase in the active cell fraction of Chloroflexota in incubations with thiosulfate was almost equivalent to the activity increase in glucose treatments (Fig. 4). This result was rather surprising, as the majority of Chloroflexota ASVs in both samples were not related to known thiotrophs. Almost all Chlorofexota ASVs in the Bizovac sample belonged to the clade SBR1031 and the family Anaerolineaceae. These were previously found to occur in microbial mats at temperatures of 25–60°C in hot springs of the Tibetan Plateau [117], Costa Rica [118], and Saudi Arabia [119]. Most members of the Anaerolineaceae are known to be mesophilic or thermophilic chemoheterotrophs that grow mainly under anaerobic conditions and metabolize sugars fermentatively [120]. In addition to fermentative sugar metabolism, SBR1031 members also encode key genes for acetogenic dehydrogenation [121]. Thus, the stimulation of Chloroflexota by thiosulfate amendments observed here was unexpected. It is possible that either members of Chloroflexota with yet undescribed metabolism are present in the biofilm samples, or that despite the relatively short incubation time, we are observing the results of microbial interactions in which compounds produced and released by thiosulfate oxidizers, such as the thiotrophic gammaproteobacterial community, actually stimulated Chloroflexota. On the other hand, Chloroflexota ASVs from the initial Tuhelj biofilms, apart from the Anaerolineaceae family, mostly belonged to the Chloroflexaceae family, known to grow photoautotrophically, suggesting an ecological role as primary producers in hot spring environments [122], but also previously shown to utilize acetate [123]. Still, the acetate amendment in this biofilm resulted in slightly reduced activity of Chloroflexota, contrary to our expectations.
Conclusion
In this study, we for the first time explored the microbial community composition of biofilms from geothermal springs and wells in northern and north-eastern Croatia. We showed that biofilm microbial communities display a stable and site-specific composition over three sampling seasons, and determined that temperature and spring source had the greatest influence on biofilm community composition, while all other tested physicochemical factors showed only minor influence. Based on this biofilm diversity screening, two geographically distant and geochemically distinct hot springs, hosting biofilms with very dissimilar microbial communities, were selected for further activity-based analyses. The here utilized method, BONCAT–CARD-FISH, has proven to be a rapid and comparatively simple way to investigate the ecophysiology of geothermal biofilm communities, allowing metabolic and taxonomic screening not constrained by cultivation limitations. However, although we demonstrated chemoorganotrophic and thiosulfate-dependent chemolithotrophic activity in both biofilms, BONCAT–CARD-FISH cannot directly link substrate uptake to microbial activity due to likely secondary interactions of microorganisms. Many results obtained in the performed activity measurements contradicted our hypothesis on dominant metabolism and likely involved biofilm members that we had derived from hot spring geochemistry and 16S rRNA gene amplicon community profiling. This outcome emphasizes the necessity for further physiology- and activity-targeted experimentation to fully understand microbial interactions and metabolism, even within the relatively well-defined habitat of hot spring biofilms.
Data Availability
16S rRNA gene amplicon sequencing data have been deposited in SRA (the Sequence Read Archive) under the BioProject accession number PRJNA889237.
References
Limberger J, Boxem T, Pluymaekers M et al (2018) Geothermal energy in deep aquifers: a global assessment of the resource base for direct heat utilization. Renew Sustain Energy Rev 82:961–975. https://doi.org/10.1016/j.rser.2017.09.084
Barbier E (2002) Geothermal energy technology and current status: an overview. Renew Sustain Energy Rev 6:3–65. https://doi.org/10.1016/S1364-0321(02)00002-3
Podar PT, Yang Z, Björnsdóttir SH, Podar M (2020) Comparative analysis of microbial diversity across temperature gradients in hot springs from Yellowstone and Iceland. Front Microbiol 11:1–16. https://doi.org/10.3389/fmicb.2020.01625
Inskeep WP, Rusch DB, Jay ZJ et al (2010) Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PloS One 5:e9773. https://doi.org/10.1371/journal.pone.0009773
Des Marais DJ, Walter MR (2019) Terrestrial hot spring systems: introduction. Astrobiology 19:1419–1432. https://doi.org/10.1089/ast.2018.1976
Stetter KO (1996) Hyperthermophilic procaryotes. FEMS Microbiol Rev 18:149–158. https://doi.org/10.1111/j.1574-6976.1996.tb00233.x
Purcell D, Sompong U, Yim LC et al (2007) The effects of temperature, pH and sulphide on the community structure of hyperthermophilic streamers in hot springs of northern Thailand. FEMS Microbiol Ecol 60:456–466. https://doi.org/10.1111/j.1574-6941.2007.00302.x
Strazzulli A, Iacono R, Giglio R et al (2017) Metagenomics of hyperthermophilic environments: biodiversity and biotechnology. In: Chénard C, Lauro F (eds) Microbial ecology of extreme environments. Springer, Cham, pp 103–135. https://doi.org/10.1007/978-3-319-51686-8_5
Debnath T, Kujur RRA, Mitra R, Das SK (2019) Diversity of microbes in hot springs and their sustainable use. In: Satyanarayana T, Johri B, Das S (eds) Microbial diversity in ecosystem sustainability and biotechnological applications. Springer, Singapore, pp 159–186. https://doi.org/10.1007/978-981-13-8315-1_6
Hou W, Wang S, Dong H et al (2013) A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan Province China Using 16S rRNA Gene Pyrosequencing. PLoS One 8:e53350. https://doi.org/10.1371/journal.pone.0053350
Wemheuer B, Taube R, Akyol P et al (2013) Microbial diversity and biochemical potential encoded by thermal spring metagenomes derived from the Kamchatka peninsula. Archaea 2013:136714. https://doi.org/10.1155/2013/136714
Li L, Ma Z (2019) Global microbiome diversity scaling in hot springs with DAR (diversity-area relationship) profiles. Front Microbiol 10:118. https://doi.org/10.3389/fmicb.2019.00118
Sharp CE, Brady AL, Sharp GH et al (2014) Humboldt’s spa: microbial diversity is controlled by temperature in geothermal environments. ISME J 8:1166–1174. https://doi.org/10.1038/ismej.2013.237
Nishida A, Thiel V, Nakagawa M et al (2018) Effect of light wavelength on hot spring microbial mat biodiversity. PLoS One 13:e0191650. https://doi.org/10.1371/journal.pone.0191650
D'Imperio S, Lehr CR, Breary M, McDermott TR (2007) Autecology of an arsenite chemolithotroph: sulfide constraints on function and distribution in a geothermal spring. Appl Env Micorbiol 73:7067–7074. https://doi.org/10.1128/AEM.01161-07
Hamilton TL, Bennett AC, Murugapiran SK, Havig JR (2019) Anoxygenic phototrophs span geochemical gradients and diverse morphologies in terrestrial geothermal springs. Msystems 4:e00498–e00419. https://doi.org/10.1128/mSystems.00498-19
Houghton KM, Carere CR, Stott MB, McDonald IR (2019) Thermophilic methanotrophs: in hot pursuit. FEMS Microbiol Ecol 95:fiz125. https://doi.org/10.1093/femsec/fiz125
DeCastro ME, Rodríguez-Belmonte E, González-Siso MI (2016) Metagenomics of thermophiles with a focus on discovery of novel thermozymes. Front Microbiol 7:1–21. https://doi.org/10.3389/fmicb.2016.01521
Brock TD, Freeze H (1969) Thermus aquaticus gen. n. and sp. n., a non-sporulating extreme thermophile. J Bacteriol 98:289–297
Chien A, Edgar DB, Trela JM (1976) Deoxyribonucleic acid polymerase from the extreme thermophile Thermus aquaticus. J Bacteriol 127:1550–1557. https://doi.org/10.1128/jb.127.3.1550-1557.1976
Schuler CG, Havig JR, Hamilton TL (2017) Hot spring microbial community composition, morphology, and carbon fixation: implications for interpreting the ancient rock record. Front Earth Sci 5:1–17. https://doi.org/10.3389/feart.2017.00097
Ward DM, Weller R, Shiea J, Castenholz RW, Cohen Y (1989) Hot spring microbial mats: anoxygenic and oxygenic mats of possible evolutionary significance. In: Cohen Y, Rosenburg E (eds) Microbial mats: physiological ecology of benthic microbial communities. American Society for Microbiology, Washington, DC, pp 3–15
Donlan RM, Costerton JW (2002) Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. https://doi.org/10.1128/CMR.15.2.167-193.2002
Prieto-Barajas CM, Valencia-Cantero E, Santoyo G (2018) Microbial mat ecosystems: structure types, functional diversity, and biotechnological application. Electron J Biotechnol 31:48–56. https://doi.org/10.1016/j.ejbt.2017.11.001
Costerton JW (1995) Overview of microbial biofilms. J Ind Microbiol 15:137–140. https://doi.org/10.1007/BF01569816
Costerton JW, Lewandowski Z, Caldwell DE et al (1995) Microbial biofilms. Annu Rev Microbiol 49:711–745. https://doi.org/10.1146/annurev.mi.49.100195.003431
Marić S, Vraneš J (2007) Characteristics and significance of microbial biofilm formation. Period Biol 109:115–121
Msarah MJ, Faiz M, Yusoff M et al (2018) Extreme environment: biofilms and microbial diversity. Malays J Microbiol 14:435–443
Kour D, Rana KL, Kaur T et al (2019) Microbial biofilms: functional annotation and potential applications in agriculture and allied sectors. In: Singh BP (ed) Yadav MK. Elsevier, New and future developments in microbial biotechnology and bioengineering, pp 283–301. https://doi.org/10.1016/B978-0-444-64279-0.00018-9
Rana KL, Kour D, Yadav AN et al (2019) Agriculturally important microbial biofilms: biodiversity, ecological significances, and biotechnological applications. In: Singh B (ed) Yadav MK. Elsevier, New and future developments in microbial biotechnology and bioengineering, pp 221–265. https://doi.org/10.1016/B978-0-444-64279-0.00016-5
Dobretsov S, Abed RMM, Al Maskari SM et al (2011) Cyanobacterial mats from hot springs produce antimicrobial compounds and quorum-sensing inhibitors under natural conditions. J Appl Phycol 23:983–993. https://doi.org/10.1007/s10811-010-9627-2
Tripathi C, Mahat NK, Rani P et al (2016) Draft genome sequence of Lampropedia cohaerens strain CT6 T isolated from arsenic rich microbial mats of a Himalayan hot water spring. Stand Genomic Sci 11:1–7. https://doi.org/10.1186/s40793-016-0179-1
Ward DM, Cohan FM (2005) Microbial diversity in hot spring cyanobacterial mats: pattern and prediction. In Geothermal biology and geochemistry in Yellowstone National Park: Proceedings of the Thermal Biology Institute Workshop. https://digitalcollections.wesleyan.edu/object/biolfp-75 Accessed 15 February 2023
Swingley WD, Meyer-Dombard DR, Shock EL et al (2012) Coordinating environmental genomics and geochemistry reveals metabolic transitions in a hot spring ecosystem. PLoS One 7:e38108. https://doi.org/10.1371/JOURNAL.PONE.0038108
Schubotz F, Meyer-Dombard DR, Bradley AS et al (2013) Spatial and temporal variability of biomarkers and microbial diversity reveal metabolic and community flexibility in streamer biofilm communities in the Lower Geyser Basin, Yellowstone National Park. Geobiology 11:549–569. https://doi.org/10.1111/gbi.12051
Venturi S, Crognale S, di Benedetto F et al (2022) Interplay between abiotic and microbial biofilm-mediated processes for travertine formation: insights from a thermal spring (Piscine Carletti, Viterbo, Italy). Geobiology 20:837–856. https://doi.org/10.1111/gbi.12516
Møller S, Sternberg C, Andersen JB et al (1998) In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl Environ Microbiol 64:721–732. https://doi.org/10.1128/aem.64.2.721-732.1998
Weiland-Bräuer N (2021) Friends or foes—microbial interactions in nature. Biology 10:496. https://doi.org/10.3390/biology10060496
Braissant O, Astasov-Frauenhoffer M, Waltimo T, Bonkat G (2020) Review of methods to determine viability, vitality, and metabolic rates in microbiology. Front Microbiol 11:547458. https://doi.org/10.3389/fmicb.2020.547458
Schubotz F, Hays LE, Meyer-Dombard DAR et al (2015) Stable isotope labeling confirms mixotrophic nature of streamer biofilm communities at alkaline hot springs. Front Microbiol 6:42. https://doi.org/10.3389/fmicb.2015.00042
Wagner M (2009) Single-cell ecophysiology of microbes as revealed by Raman microspectroscopy or secondary ion mass spectrometry imaging. Ann Rev Microbiol 63:411–429. https://doi.org/10.1146/annurev.micro.091208.073233
Hatzenpichler R, Krukenberg V, Spietz RL, Jay ZJ (2020) Next generation physiology approaches to study microbiome function at single cell level. Nat Rev Microbiol 18:241–256. https://doi.org/10.1038/s41579-020-0323-1
Alcolombri U, Pioli R, Stocker R, Berry D (2022) Single-cell stable isotope probing in microbial ecology. ISME Comm 2:1–9. https://doi.org/10.1038/s43705-022-00142-3
Azeredo J, Azevedo NF, Briandet R et al (2017) Critical review on biofilm methods. Crit Rev Microbiol 43:313–351. https://doi.org/10.1080/1040841X.2016.1208146
Hatzenpichler R, Scheller S, Tavormina PL et al (2014) In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry. Environ Microbiol 16:2568–2590. https://doi.org/10.1111/1462-2920.12436
Kindaichi T, Ito T, Okabe S (2004) Ecophysiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms as determined by microautoradiography-fluorescence in situ hybridization. Appl Environ Microbiol 70:1641–1650. https://doi.org/10.1128/AEM.70.3.1641-1650.2004
Musat N, Musat F, Weber PK, Pett-Ridge J (2016) Tracking microbial interactions with NanoSIMS. Curr OBiotech 41:114–121. https://doi.org/10.1016/j.copbio.2016.06.007
Brock TD (1969) Vertical zonation in hot spring algal mats. Phycologia 8:201–205. https://doi.org/10.2216/i0031-8884-8-3-201.1
Brock TD, Brock ML (1969) Effect of light intensity on photosynthesis by thermal algae adapted to natural and reduced sunlight. Limnol Oceanogr 14:334–341. https://doi.org/10.4319/lo.1969.14.3.0334
Bauld J, Brock TD (1973) Ecological studies of Chloroflexis, a gliding photosynthetic bacterium. Archiv für Mikrobiologie 92:267–284. https://doi.org/10.1007/BF00409281
Doemel WN, Brock TD (1977) Structure, growth, and decomposition of laminated algal-bacterial mats in alkaline hot springs. Appl Env Microbiol 34:433–452. https://doi.org/10.1128/aem.34.4.433-452.1977
Leizeaga A, Estrany M, Forn I, Sebastián M (2017) Using click-chemistry for visualizing in situ changes of translational activity in planktonic marine bacteria. Front Microbiol 8:2360. https://doi.org/10.3389/fmicb.2017.02360
Nuñez J, Renslow R, Cliff JB, Anderton CR (2018) NanoSIMS for biological applications: current practices and analyses. Biointerphases 13:03B301. https://doi.org/10.1116/1.4993628
Renslow RS, Lindemann SR, Cole JK et al (2016) Quantifying element incorporation in multispecies biofilms using nanoscale secondary ion mass spectrometry image analysis. Biointerphases 11:02A322. https://doi.org/10.1116/1.4941764
Hatzenpichler R, Connon SA, Goudeau D et al (2016) Visualizing in situ translational activity for identifying and sorting slow-growing archaeal–bacterial consortia. PNAS 113:4069–4078. https://doi.org/10.1073/pnas.1603757113
Samo TJ, Smriga S, Malfatti F et al (2014) Broad distribution and high proportion of protein synthesis active marine bacteria revealed by click chemistry at the single cell level. Front Mar Sci 1:48. https://doi.org/10.3389/fmars.2014.00048
Hatzenpichler R, Orphan VJ (2015) Detection of protein-synthesizing microorganisms in the environment via bioorthogonal noncanonical amino acid tagging (BONCAT). In: McGenity TJ, Timmis KN, Nogales B (eds) Hydrocarbon and lipid microbiology protocols. Springer, Berlin, pp 145–157. https://doi.org/10.1007/8623_2015_61
Landgraf P, Antileo ER, Schuman EM, Dieterich DC (2015) BONCAT: metabolic labeling, click chemistry, and affinity purification of newly synthesized proteomes. Methods Mol Biol 1266:199–215. https://doi.org/10.1007/978-1-4939-2272-7_14
Lindivat M, Larsen A, Hess-Erga OK et al (2020) Bioorthogonal non-canonical amino acid tagging combined with flow cytometry for determination of activity in aquatic microorganisms. Front Microbiol 11:1929. https://doi.org/10.3389/fmicb.2020.01929
Steward KF, Eilers B, Tripet B et al (2020) Metabolic implications of using BioOrthogonal Non-Canonical Amino Acid Tagging (BONCAT) for tracking protein synthesis. Front Microbiol 11:197. https://doi.org/10.3389/fmicb.2020.00197
Babin BM, Atangcho L, van Eldijk MB et al (2017) Selective proteomic analysis of antibiotic-tolerant cellular subpopulations in Pseudomonas aeruginosa biofilms. mBio 8:e01593-17. https://doi.org/10.1128/MBIO.01593-17
Marković T, Sladović Ž, Domitrović D et al (2022) Current utilization and hydrochemical characteristics of geothermal aquifers in the Bjelovar sub-depression. Geol Croat 75:223–233. https://doi.org/10.4154/gc.2022.21
Mitrović M, Kostešić E, Marković T et al (2022) Microbial community composition and hydrochemistry of underexplored geothermal waters in Croatia. Syst Appl Microbiol 45:126359. https://doi.org/10.1016/J.SYAPM.2022.126359
Monier JM, Lindow SE (2003) Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. PNAS 26:15977–15982. https://doi.org/10.1073/pnas.2436560100
Berlanga M, Guerrero R (2016) Living together in biofilms: the microbial cell factory and its biotechnological implications. Microb Cell Fact 15:1–10. https://doi.org/10.1186/s12934-016-0569-5
Parada AE, Needham DM, Fuhrman JA (2016) Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. https://doi.org/10.1111/1462-2920.13023
Apprill A, Mcnally S, Parsons R, Weber L (2015) Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137. https://doi.org/10.3354/ame01753
Pjevac P, Hausmann B, Schwarz J et al (2021) An economical and flexible dual barcoding, two-step PCR approach for highly multiplexed amplicon sequencing. Front Microbiol 12:669776. https://doi.org/10.3389/fmicb.2021.669776
Bushnell B (2014) BBMap: a fast, accurate, splice-aware aligner. in, 1–2. Available at: https://escholarship.org/uc/item/1h3515gn . Accessed October 13 2022
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016a) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/NMETH.3869
Callahan BJ, Sankaran K, Fukuyama JA et al (2016b) Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Res 5:1492. https://doi.org/10.12688/F1000RESEARCH.8986.2
Pruesse E, Peplies J, Glöckner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823. https://doi.org/10.1093/BIOINFORMATICS/BTS252
Huang R, Soneson C, Ernst FG et al (2021) TreeSummarizedExperiment: a S4 class for data with hierarchical structure. F1000Res:9. https://doi.org/10.12688/f1000research.26669.2
McMurdie PJ, Holmes S (2013) phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. https://doi.org/10.1371/JOURNAL.PONE.0061217
Ernst F, Shetty S, Borman T, Lahti L (2022) mia: Microbiome analysis. R package version 1.4.0., https://github.com/microbiome/mia. Available at: https://www.bioconductor.org/packages/release/bioc/html/mia.html Accessed October 13, 2022
Barnett DJ, Arts IC, Penders J (2021) microViz: an R package for microbiome data visualization and statistics. J Open Source Softw 6:3201. https://doi.org/10.21105/JOSS.03201
Wickham H (2009) ggplot2: elegant graphics for data analysis. Springer-Verlag, New York. https://doi.org/10.1007/978-0-387-98141-3
Kappler U, Dahl C (2001) Enzymology and molecular biology of prokaryotic sulfite oxidation. FEMS Microbiol Lett 203:1–9. https://doi.org/10.1111/J.1574-6968.2001.TB10813.X
Thiel K, Vuorio E, Aro EM, Kallio PT (2017) The effect of enhanced acetate influx on Synechocystis sp. PCC 6803 metabolism. Microb Cell Fact 16:21. https://doi.org/10.1186/S12934-017-0640-X
Chuang DSW, Liao JC (2021) Role of cyanobacterial phosphoketolase in energy regulation and glucose secretion under dark anaerobic and osmotic stress conditions. Metab Eng 65:255–262. https://doi.org/10.1016/J.YMBEN.2020.12.004
Hidayat MY, Saud HM, Samsudin AA (2017) Isolation and characterisation of sulphur oxidizing bacteria isolated from hot spring in Malaysia for biological deodorisation of hydrogen sulphide in chicken manure. Media Peternakan 40:178–187. https://doi.org/10.5398/medpet.2017.40.3.178
Reichart NJ, Jay ZJ, Hatzenpichler R et al (2020) Activity-based cell sorting reveals responses of uncultured archaea and bacteria to substrate amendment. ISME J 14:2851–2861. https://doi.org/10.1038/s41396-020-00749-1
Pin L, Eiler A, Fazi S, Friberg N (2021) Two different approaches of microbial community structure characterization in riverine epilithic biofilms under multiple stressors conditions: developing molecular indicators. Mol Ecol Resour 21:1200–1215. https://doi.org/10.1111/1755-0998.13341
Schmidt H, Eickhorst T, Tippkötter R (2012) Evaluation of tyramide solutions for an improved detection and enumeration of single microbial cells in soil by CARD-FISH. J Microbiol Methods 91:399–405. https://doi.org/10.1016/J.MIMET.2012.09.021
Kubota K (2013) CARD-FISH for environmental microorganisms: technical advancement and future applications. Microbes Environ 28:3–12. https://doi.org/10.1264/jsme2.ME12107
Daims H, Lücker S, Wagner M (2006) daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol 8:200–213. https://doi.org/10.1111/j.1462-2920.2005.00880.x
Sompong U, Hawkins PR, Besley C, Peerapornpisal Y (2005) The distribution of cyanobacteria across physical and chemical gradients in hot springs in Northern Thailand. FEMS Microb Ecol 52:365–376. https://doi.org/10.1016/j.femsec.2004.12.007
Power JF, Carere CR, Lee CK et al (2018) Microbial biogeography of 925 geothermal springs in New Zealand. Nat Commun 9:2876. https://doi.org/10.1038/s41467-018-05020-y
Portillo MC, Sririn V, Kanoksilapatham W, Gonzalez JM (2009) Differential microbial communities in hot spring mats from Western Thailand. Extremophiles 13:321–331. https://doi.org/10.1007/s00792-008-0219-x
Huang Q, Dong CZ, Dong RM et al (2011) Archaeal and bacterial diversity in hot springs on the Tibetan Plateau, China. Extremophiles 15:549–563. https://doi.org/10.1007/s00792-011-0386-z
Coman C, Chiriac CM, Robeson MS et al (2015) Structure, mineralogy, and microbial diversity of geothermal spring microbialites associated with a deep oil drilling in Romania. Front Microbiol 6:1–14. https://doi.org/10.3389/fmicb.2015.00253
Coman C, Drugă B, Hegedus A et al (2013) Archaeal and bacterial diversity in two hot spring microbial mats from a geothermal region in Romania. Extremophiles 17:523–534. https://doi.org/10.1007/s00792-013-0537-5
Alcamán-Arias ME, Pedrós-Alió C, Tamames J et al (2018) Diurnal changes in active carbon and nitrogen pathways along the temperature gradient in porcelana hot spring microbial mat. Front Microbiol 9:1–17. https://doi.org/10.3389/fmicb.2018.02353
Mackenzie R, Pedrós-Alió C, Díez B (2013) Bacterial composition of microbial mats in hot springs in Northern Patagonia: variations with seasons and temperature. Extremophiles 17:123–136. https://doi.org/10.1007/s00792-012-0499-z
Guo L, Wang G, Sheng Y et al (2020) Temperature governs the distribution of hot spring microbial community in three hydrothermal fields, Eastern Tibetan Plateau Geothermal Belt, Western China. Sci Total Environ 720:137574. https://doi.org/10.1016/j.scitotenv.2020.137574
Toshchakov SV, Izotova AO, Vinogradova EN et al (2021) Culture-independent survey of thermophilic microbial communities of the north Caucasus. Biology 10:1352. https://doi.org/10.3390/biology10121352
Rozanov AS, Bryanskaya AV, Ivanisenko TV et al (2017) Biodiversity of the microbial mat of the Garga hot spring. BMC Evol Biol 17:37–49. https://doi.org/10.1186/s12862-017-1106-9
Della Porta G, Hoppert M, Hallmann C et al (2022) The influence of microbial mats on travertine precipitation in active hydrothermal systems (Central Italy). Depositional Rec 8:165–209. https://doi.org/10.1002/dep2.147
Valeriani F, Crognale S, Protano C et al (2018) Metagenomic analysis of bacterial community in a travertine depositing hot spring. New Microbiol 41:126–135
Wasmund K, Mußmann M, Loy A (2017) The life sulfuric: microbial ecology of sulfur cycling in marine sediments. Environ Microbiol Rep 9:323–344. https://doi.org/10.1111/1758-2229.12538
Chernitsyna S, Elovskaya I, Pogodaeva T et al (2023) Bacterial communities in a gradient of abiotic factors near a sulfide thermal spring in Northern Baikal. Diversity 15:298. https://doi.org/10.3390/d15020298
Ghilamicael AM, Budambula NLM, Anami SE et al (2017) Evaluation of prokaryotic diversity of five hot springs in Eritrea. BMC Microbiol 17:1–13. https://doi.org/10.1186/s12866-017-1113-4
Gao S, Li Z, Hou Y et al (2022) Effects of different carbon sources on the efficiency of sulfur-oxidizing denitrifying microorganisms. Environ Res 204:111946. https://doi.org/10.1016/J.ENVRES.2021.111946
Zhang Y, Sun L, Zhou J (2019) Simultaneous biological and chemical removal of sulfate and Fe(II)EDTA-NO in anaerobic conditions and regulation of sulfate reduction products. Minerals 9:330. https://doi.org/10.3390/min9060330
Li W, Lin J, Zhang L et al (2016) Mixotrophic denitrification desulfurization wastewater treatment process: bioreactor performance and analysis of the microbial community. Pol J Environ Stud 25:2491–2497. https://doi.org/10.15244/pjoes/63335
Liu Q, Huang C, Chen X et al (2020) Succession of functional bacteria in a denitrification desulphurisation system under mixotrophic conditions. Environ Res 188:109708. https://doi.org/10.1016/j.envres.2020.109708
Xu XJ, Chen C, Wang AJ et al (2014) Bioreactor performance and functional gene analysis of microbial community in a limited-oxygen fed bioreactor for co-reduction of sulfate and nitrate with high organic input. J Hazard Mater 278:250–257. https://doi.org/10.1016/j.jhazmat.2014.06.006
Lee DJ, Wong BT, Adav SS (2014) Azoarcus taiwanensis sp nov, a denitrifying species isolated from a hot spring. Appl Microbiol Biotechnol 98:1301–1307. https://doi.org/10.1007/s00253-013-4976-9
Boden R, Hutt LP, Rae AW (2017) Reclassification of Thiobacillus aquaesulis (Wood & Kelly, 1995) as Annwoodia aquaesulis gen. nov., comb. nov., transfer of Thiobacillus (Beijerinck, 1904) from the Hydrogenophilales to the Nitrosomonadales, proposal of Hydrogenophilalia class. nov. within the “Proteobacteria”, and four new families within the orders Nitrosomonadales and Rhodocyclales. Int J Syst Evol Microbiol 67:1191–1205
Kawai S, Ishikawa M, Hanada S, Haruta S (2022) Hydrogenophilus thiooxidans sp. nov., a moderately thermophilic chemotrophic bacterium unable to grow on hydrogen gas, isolated from hot spring microbial mats. Int J Syst Evol Microbiol 72:005355. https://doi.org/10.1099/ijsem.0.005355
Mori K, Suzuki KI (2008) Thiofaba tepidiphila gen. nov., sp. nov., a novel obligately chemolithoautotrophic, sulfur-oxidizing bacterium of the Gammaproteobacteria isolated from a hot spring. Int J Syst Evol Microbiol 58:1885–1891. https://doi.org/10.1099/ijs.0.65754-0
Singh T, Kshirsagar PR, Das A et al (2019) Implications of microbial thiosulfate utilization in red clay sediments of the Central Indian Basin: the Martian analogy. Geochem Geophys Geosyst 20:708–729. https://doi.org/10.1029/2018GC007640
Gutierrez T (2019) Aerobic hydrocarbon-degrading Gammaproteobacteria: Porticoccus. In: McGenity T (ed) Taxonomy, genomics and ecophysiology of hydrocarbon-degrading microbes. Handbook of Hydrocarbon and Lipid Microbiology. Springer, Cham. https://doi.org/10.1007/978-3-030-14796-9_32
Dyksma S, Lenk S, Sawicka JE, Mußmann M (2018) Uncultured Gammaproteobacteria and Desulfobacteraceae account for major acetate assimilation in a coastal marine sediment. Front Microbiol 9:3124. https://doi.org/10.3389/fmicb.2018.03124
Winkel M, Pjevac P, Kleiner M et al (2014) Identification and activity of acetate-assimilating bacteria in diffuse fluids venting from two deep-sea hydrothermal systems. FEMS Microbiol Ecol 90:731–746. https://doi.org/10.1111/1574-6941.12429
Kutscha R, Pflügl S (2020) Microbial upgrading of acetate into value-added products—examining microbial diversity, bioenergetic constraints and metabolic engineering approaches. Int J Mol Sci 21:1–30. https://doi.org/10.3390/ijms21228777
Wang S, Hou W, Dong H et al (2013) Control of temperature on microbial community structure in hot springs of the Tibetan Plateau. PLoS One 8:e62901. https://doi.org/10.1371/journal.pone.0062901
Uribe-Lorío L, Brenes-Guillén L, Hernández-Ascencio W et al (2019) The influence of temperature and pH on bacterial community composition of microbial mats in hot springs from Costa Rica. MicrobiologyOpen 8:1–26. https://doi.org/10.1002/mbo3.893
Yasir M, Qureshi AK, Srinivasan S et al (2020) Domination of filamentous anoxygenic phototrophic bacteria and prediction of metabolic pathways in microbial mats from the hot springs of Al Aridhah. Folia Biol 66:24–35
Yamada T, Sekiguchi Y (2018) A naerolineaceae. BMSAB 9:1–5. https://doi.org/10.1002/9781118960608.fbm00301
Xia Y, Wang Y, Wang Y et al (2016) Cellular adhesiveness and cellulolytic capacity in Anaerolineae revealed by omics-based genome interpretation. Biotechnol Biofuels 9:1–13. https://doi.org/10.1186/s13068-016-0524-z
Gupta RS, Chander P, George S (2013) Phylogenetic framework and molecular signatures for the class Chloroflexi and its different clades; proposal for division of the class Chloroflexi class. nov. into the suborder Chloroflexineae subord. nov., consisting of the emended family Oscillochlorida. Antonie Van Leeuwenhoek 103:99–119. https://doi.org/10.1007/s10482-012-9790-3
Nübel U, Bateson MM, Vandieken V et al (2002) Microscopic examination of distribution and phenotypic properties of phylogenetically diverse Chloroflexaceae-related bacteria in hot spring microbial mats. Appl Environ Microbiol 68:4593–4603. https://doi.org/10.1128/AEM.68.9.4593-4603.2002
Acknowledgements
The authors thank Jasmin Schwarz and Gudrun Kohl for amplicon preparation, Dr. Stephanie A. Eichorst for her invaluable advice on the CARD-FISH procedure, Prof. Dr. Holger Daims for help with daime analysis, and employees from thermal spas for providing significant information for our research.
Funding
Open access funding provided by University of Vienna. This research was funded by the Croatian Science Foundation and the European Social Fund under the Cooperation Program with Croatian scientists in the diaspora (“SCIENTIFIC COOPERATION”—Operational program “Effective Human Resources” 2014–2020; grant number PZS-2019-02-7373).
Author information
Authors and Affiliations
Contributions
EK, PP, and SO conceptualized and designed the study. EK, MM, and TM collected the samples. TM performed physiochemical analysis. EK developed the experimental setup and manipulations. EK, KK, BH, and performed formal data analysis. EK, SO, and PP wrote, reviewed, and edited the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kostešić, E., Mitrović, M., Kajan, K. et al. Microbial Diversity and Activity of Biofilms from Geothermal Springs in Croatia. Microb Ecol 86, 2305–2319 (2023). https://doi.org/10.1007/s00248-023-02239-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-023-02239-1