Abstract
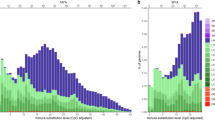
Transposable elements (TEs) are nearly ubiquitous among eukaryotic genomes, but TE contents vary dramatically among phylogenetic lineages. Several mechanisms have been proposed as drivers of TE dynamics in genomes, including the fixation/loss of a particular TE insertion by selection or drift as well as structural changes in the genome due to mutation (e.g., recombination). In particular, recombination can have a significant and directional effect on the genomic TE landscape. For example, ectopic recombination removes internal regions of long terminal repeat retrotransposons (LTR-RTs) as well as one long terminal repeat (LTR), resulting in a solo LTR. In this study, we focus on the intra-species dynamics of LTR-RTs and solo LTRs in bird genomes. The distribution of LTR-RTs and solo LTRs in birds is intriguing because avian recombination rates vary widely within a given genome. We used published linkage maps and whole genome assemblies to study the relationship between recombination rates and LTR-removal events in the chicken and zebra finch. We hypothesized that regions with low recombination rates would harbor more full-length LTR-RTs (and fewer solo LTRs) than regions with high recombination rates. We tested this hypothesis by comparing the ratio of full-length LTR-RTs and solo LTRs across chromosomes, across non-overlapping megabase windows, and across physical features (i.e., centromeres and telomeres). The chicken data statistically supported the hypothesis that recombination rates are inversely correlated with the ratio of full-length to solo LTRs at both the chromosome level and in 1-Mb non-overlapping windows. We also found that the ratio of full-length to solo LTRs near chicken telomeres was significantly lower than those ratios near centromeres. Our results suggest a potential role of ectopic recombination in shaping the chicken LTR-RT genomic landscape.




Similar content being viewed by others
References
Backström N, Forstmeier W, Schielzeth H et al (2010) The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res 20:485–495. doi:10.1101/gr.101410.109
Baker BS, Carpenterl ATC, Carolina N et al (1976) The genetic control of meiosis. Annu Rev Genet 10:53–134. doi:10.1146/annurev.ge.10.120176.000413
Barrón MG, Fiston-Lavier A-S, Petrov DA, González J (2014) Population genomics of transposable elements in Drosophila. Annu Rev Genet 48:561–581. doi:10.1146/annurev-genet-120213-092359
Benkel B, Rutherford K (2014) Endogenous avian leukosis viral loci in the Red Jungle Fowl genome assembly. Poult Sci 93:2988–2990. doi:10.3382/ps.2014-04309
Bennetzen JL, Kellogg EA (1997) Do plants have a oe-way ticket to genomic obesity? Plant Cell 9:1509–1514
Bergero R, Charlesworth D (2008) The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol 24:94–102. doi:10.1016/j.tree.2008.09.010
Braun EL, Kimball RT, Han K-L et al (2011) Homoplastic microinversions and the avian tree of life. BMC Evol Biol 11:141. doi:10.1186/1471-2148-11-141
Burt DW (2002) Origin and evolution of avian microchromosomes. Cytogenet Genome Res 96:97–112
Champely S (2015) pwr: basic functions for power analysis. R package version 1.1-3. http://CRAN.R-project.org/package=pwr
Charlesworth B (1991) Transposable elements in natural populations with a mixture of selected and neutral insertion sites. Genet Res 57:127–134. doi:10.1017/S0016672300029190
Charlesworth B, Campos JL (2014) The relations between recombination rate and patterns of molecular variation and evolution in Drosophila. Annu Rev Genet 48:383–403. doi:10.1146/annurev-genet-120213-092525
Charlesworth B, Charlesworth D (1983) The population dynamics of transposable elements. Genet Res 42:1–27. doi:10.1017/S0016672300021455
Cui J, Zhao W, Huang Z et al (2014) Low frequency of paleoviral infiltration across the avian phylogeny. Genome Biol 15:539. doi:10.1186/s13059-014-0539-3
Dolgin ES, Charlesworth B, Cutter AD (2008) Population frequencies of transposable elements in selfing and outcrossing Caenorhabditis nematodes. Genet Res (Camb) 90:317–329. doi:10.1017/S0016672308009440
Du J, Tian Z, Hans CS et al (2010) Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J 63:584–598. doi:10.1111/j.1365-313X.2010.04263.x
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14:755–763
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi:10.1093/nar/gkh340
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi:10.1093/bioinformatics/btq461
Elferink MG, van As P, Veenendaal T et al (2010) Regional differences in recombination hotspots between two chicken populations. BMC Genet 11:11. doi:10.1186/1471-2156-11-11
Ellegren H (2010) Evolutionary stasis: the stable chromosomes of birds. Trends Ecol Evol 25:283–291. doi:10.1016/j.tree.2009.12.004
Ellinghaus D, Kurtz S, Willhoeft U (2008) LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform 9:18. doi:10.1186/1471-2105-9-18
Epplen JT, Leipoldt M, Engel W, Schmidtke J (1978) DNA sequence organisation in avian genomes. Chromosoma 69:307–321. doi:10.1007/BF00332134
Farré M, Micheletti D, Ruiz-Herrera A (2013) Recombination rates and genomic shuffling in human and chimpanzee—a new twist in the chromosomal speciation theory. Mol Biol Evol 30:853–864. doi:10.1093/molbev/mss272
Frahry MB, Sun C, Chong RA, Mueller RL (2015) Low levels of LTR retrotransposon deletion by ectopic recombination in the gigantic genomes of salamanders. J Mol Evol 80:120–129. doi:10.1007/s00239-014-9663-7
Fridolfsson A-K, Heng H, Copeland NG et al (1998) Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc Natl Acad Sci USA 95:8147–8152
González J, Petrov DA (2012) Evolutionary of genome content: population dynamics of transposable elements in flies and humans. In: Anisimova M (ed) Evolutionary genomics: statistical and computational methods, vol 1. Springer Humana, Totowa, NJ, pp 361–383
Goodchild NL, Wilkinson DA, Mager DL (1993) Recent evolutionary expansion of a subfamily of RTVL-H Human Endogenous Retrovirus-like elements. Virology 196:778–788
Gregory TR (2002) A bird’s-eye view of the C-value enigma: genome size, cell size, and metabolic rate in the class Aves. Evolution (NY) 56:121–130
Gregory TR, Andrews CB, McGuire JA, Witt CC (2009) The smallest avian genomes are found in hummingbirds. Proc R Soc B Biol Sci 276:3753–3757. doi:10.1098/rspb.2009.1004
Groenen MAM, Wahlberg P, Foglio M et al (2009) A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Res 19:510–519. doi:10.1101/gr.086538.108
Hayward A, Grabherr M, Jern P (2013) Broad-scale phylogenomics provides insights into retrovirus-host evolution. Proc Natl Acad Sci USA 110:20146–20151. doi:10.1073/pnas.1315419110
Hill WG, Robertson A (1966) The effect of linkage on limits to artificial selection. Genet Res 8:269–294. doi:10.1017/S0016672300010156
Hinrichs AS, Karolchik D, Baertsch R et al (2006) The UCSC genome browser database: update 2006. Nucl Acids Res 34:D590–D598. doi:10.1093/nar/gkj144
Huda A, Polavarapu N, Jordan IK, McDonald JF (2008) Endogenous retroviruses of the chicken genome. Biol Direct 3:9. doi:10.1186/1745-6150-3-9
Hughes AL, Hughes MK (1995) Small genomes for better flyers. Nature 377:391
ICGSC (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716. doi:10.1038/nature03154
Jurka J, Kapitonov VV, Pavlicek A et al (2005) Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110:462–467. doi:10.1159/000084979
Kaback DB, Guacci V, Barber D, Mahon JW (1992) Chromosome size-dependent control of meiotic recombination. Science 256:228–232
Kaback DB, Barber D, Mahon J et al (1999) Chromosome size-dependent control of meiotic reciprocal recombination in Saccharomyces cerevisiae: the role of crossover interference. Genetics 152:1475–1486
Katzourakis A, Pereira V, Tristem M (2007) Effects of recombination rate on human endogenous retrovirus fixation and persistence. J Virol 81:10712–10717. doi:10.1128/JVI.00410-07
Kim JM, Vanguri S, Boeke JD et al (1998) Transposable elements and genome organization : a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res 8:464–478. doi:10.1101/gr.8.5.464
Langley CH, Montgomery E, Hudson R et al (1988) On the role of unequal exchange in the containment of transposable element copy number. Genet Res 52:223–235. doi:10.1017/S0016672300027695
Lichten M, Borts RH, Haber JE (1986) Meiotic gene conversion and crossing over between dispersed homologous sequences occurs frequently in Saccharomyces cerevisiae. Genetics 115:233–246
Liu GE, Hou Y, Brown T (2013) Analysis of CR1 repeats in the zebra finch genome. Syst Cybern Inform 11:66–71
Lynch M, Conery JS (2003) The origins of genome complexity. Science 302:1401–1404. doi:10.1126/science.1089370
Nam K, Ellegren H (2012) Recombination drives vertebrate genome contraction. PLoS Genet 8:e1002680. doi:10.1371/journal.pgen.1002680
Petrov DA (2002) Mutational equilibrium model of genome size evolution. Theor Popul Biol 61:531–544. doi:10.1006/tpbi.2002.1605
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi:10.1093/bioinformatics/btq033
R Core Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16:2–3
Romanov MN, Farré M, Lithgow PE et al (2014) Reconstruction of gross avian genome structure, organization and evolution suggests that the chicken lineage most closely resembles the dinosaur avian ancestor. BMC Genom 15:1060. doi:10.1186/1471-2164-15-1060
Sanmiguel P, Tikhonov A, Jin Y et al (1996) Nested retrotransposons in the intergenic gegions of the maize genome. Science (80-)274:765–768
Sasaki M, Lange J, Keeney S (2010) Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol 11:182–195. doi:10.1038/nrm2849
Sniegowski PD, Charlesworth B (1994) Transposable element numbers in cosmopolitan inversions From a natural population of Drosophila melanogaster. Genetics 137:815–827
Sun C, Shepard DB, Chong RA et al (2012) LTR retrotransposons contribute to genomic gigantism in plethodontid salamanders. Genome Biol Evol 4:168–183. doi:10.1093/gbe/evr139
Tiley GP, Burleigh JG (2015) The relationship of recombination rate, genome structure, and patterns of molecular evolution across angiosperms. BMC Evol Biol 15:194. doi:10.1186/s12862-015-0473-3
Treangen TJ, Salzberg SL (2012) Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 13:36–46. doi:10.1038/nrg3117
Uno Y, Nishida C, Tarui H et al (2012) Inference of the protokaryotypes of amniotes and tetrapods and the evolutionary processes of microchromosomes from comparative gene mapping. PLoS One 7:2–13. doi:10.1371/journal.pone.0053027
Vitte C, Panaud O (2003) Formation of solo-LTRs through unequal homologous recombination counterbalances amplifications of LTR retrotransposons in rice Oryza sativa L. Mol Biol Evol 20:528–540. doi:10.1093/molbev/msg055
Waterhouse AM, Procter JB, Martin DMA et al (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi:10.1093/bioinformatics/btp033
Wicker T, Robertson JS, Schulze SR et al (2005) The repetitive landscape of the chicken genome. Genome Res 15:126–136. doi:10.1101/gr.2438005
Wicker T, Sabot F, Hua-Van A et al (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982. doi:10.1038/nrg2165
Zhang G, Li C, Li Q et al (2014) Comparative genomics reveals insights into avian genome evolution and adaptation. Science (80-)346:1311–1320. doi:10.1126/science.1251385
Zhou Q, Zhang J, Bachtrog D et al (2014) Complex evolutionary trajectories of sex chromosomes across bird taxa. Science (80-)346:1246338. doi:10.1126/science.1246338
Acknowledgments
We thank Purdue’s Department of Forestry and Natural Resources and the China Scholarship Council for funding Y.J., and Purdue’s Office of the Provost for funding to J.A.D. through the University Faculty Scholar program. We thank S. Steinbiss for maintaining LTRdigest, M. Groenen for providing the data on centromeres in chicken, M. Sundaram for statistical advice, members of DeWoody Lab, and anonymous reviewers and the editor for providing constructive criticism on an earlier version of this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ji, Y., DeWoody, J. Genomic Landscape of Long Terminal Repeat Retrotransposons (LTR-RTs) and Solo LTRs as Shaped by Ectopic Recombination in Chicken and Zebra Finch. J Mol Evol 82, 251–263 (2016). https://doi.org/10.1007/s00239-016-9741-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-016-9741-0