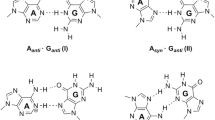
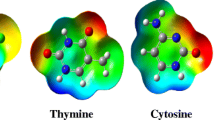
Abstract
A comparative study of the tautomeric effect of guanine (G) on stability, spectral properties, and optical response in canonical and noncanonical cytosine–guanine (C–G) base pairs has been investigated by employing density functional theory (DFT) and time-dependent DFT. Three keto-G tautomers (G1, G2, G3) have taken to interact with the C monomer of W–C type which in turn gives three C–G base pairs (C–G1, C–G2, and C–G3) with three conventional H-bonds. The geometry optimization with various ground-state properties and spectroscopic details has been estimated in the gas phase using Cam-B3LYP functional with two different basis sets 6-311++G (d, p) and Aug-cc-pVDZ at the DFT level. The W–C base pair C–G1 possesses the lowest electronic energy although G1 is not the lowest energy structure of guanine. C–G3 is found to have maximum binding and H-bond energies. Significant changes have been observed in IR and Raman spectra of C–G clusters due to the tautomeric effect of G monomer. All the absorption spectra of C–G clusters belong to the UV region, and the electronic excitations are associated with π → π* transition. The molecular orbitals associated with the electronic transitions reveal the intermolecular charge transfer characteristic of G1, G3 to C monomer. Besides this, the electrostatic potential surface plots have been used to explore the possible binding sites of isolated bases and base pairs. As the tautomeric effect influences the chemical reactions, this study can be useful for drug designing and to find the best fit between the receptors and important ligands.






Similar content being viewed by others
References
Ren XH, Wang HJ (2009) Hydrogen-bonding interaction in a complex of amino acid with N, N dimethylformamide studied by DFT calculations. J Solut Chem 38:303–313
Takano Y, Kusaka A, Nakamura Haruki (2013) Density functional study of molecular interactions in secondary structures of proteins. Biophys Soc Jpn 13:27–35
Li Y, Yang Y, Ding Y (2017) The new competitive mechanism of hydrogen bonding interactions and transition process for the hydroxyphenyl imidazo [1, 2-a] pyridine in mixed liquid solution. Sci Rep 7:1–15
Poater J, Swart M, Bickelhaupta FM, Guerra CF (2014) B-DNA structure and stability: the role of hydrogen bonding, π–π stacking interactions, twist-angle, and salvation. Org Biomol Chem 12:4691–4700
Shi Y, Jiang W, Zhangb Z, Wang Z (2017) Cooperative vibrational properties of hydrogen bonds in Watson–Crick DNA base pairs. New J Chem 41:12104–12109
Horowitz S, Trievel RC (2012) Carbon-oxygen hydrogen bonding in biological structure and function. J Biol Chem 287:41576–41582
Halder A, Data D, Seelam PP, Bhattacharyya D, Mitra A (2019) Estimating strengths of individual hydrogen bonds in RNA base pairs: toward a consensus between different computational approaches. ACS Omega 4:7354–7368
Srivastava R (2019) The role of proton transfer on mutations. Front Chem 7:1–16
Mukherjee S, Majumdar S, Bhattacharyya D (2005) Role of hydrogen bonds in protein-DNA recognition: effect of nonplanar amino groups. J. Phys Chem B 109:10484–10492
Řezáč J, Hobza P (2016) Benchmark calculations of interaction energies in noncovalent complexes and their applications. Chem Rev 116:5038–5071
Dakeshwar Kumar Verma (2018) A density functional theory (DFT) as a powerful tool for designing corrosion inhibitors in aqueous phase. IntechOpen, London
Lipkowitz Kenny B, Boyd Donald B (2002) Reviews in computational chemistry. Wiley-VCH, Hoboken
Watson JD, Crick FHC (1953) Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 171:737–738
Sÿponer J, Leszczynski J, Hobza P (1996) Structures and energies of hydrogen-bonded DNA base pairs. A nonempirical study with inclusion of electron correlation. J Phys Chem 100:1965–1974
Guerra CF, Bickelhaupt FM, Snijders JG, Baerends EJ (1999) The nature of the hydrogen bond in DNA base pairs: the role of charge transfer and resonance assistance. Chem Eur J 5:3581–3594
Guerra CF, Bickelhaupt FM, Snijders JG, Baerends EJ (2000) Hydrogen bonding in DNA base pairs: reconciliation of theory and experiment. J Am Chem Soc 122:4117–4128
Chung G, Oh H, Lee D (2005) Tautomerism and isomerism of guanine–cytosine DNA base pair: Ab initio and density functional theory approaches. J Mol Struct Theochem 730:241–249
Mons M, Dimicoli I, Piuzzi F, Tardivel B, Elhanine M (2002) Tautomerism of the DNA base guanine and its methylated derivatives as studied by gas-phase infrared and ultraviolet spectroscopy. J Phys Chem 106:5088–5094
Deepa P, Kolandaivel P (2008) Studies on tautomeric forms of guanine–cytosine base pairs of nucleic acids and their interactions with water molecules. J Biomol Struct Dyn 25:733–745
Lopes RP, Marques MPM, Valero R, Tomkinson J, Carvalho LAEB (2012) Guanine: a combined study using vibrational spectroscopy and theoretical methods. Spectrosc-Int J. 27:273–292
Heidarnezhada Z, Heidarnezhada F, Ganievb I, Obidov Z (2013) A DFT study on tautomer stability of 4-hydroxyquinoline considering solvent effect and NBO analysis. Chem Sci Trans 2:1370–1378
Babu ND, Jayaprakash D (2015) Density functional theory (DFT) studies of the stability of tautomers and equilibrium constants of cyanuric acid (CA) in different solvents. J Chem Pharm Res 7:1155–1160
Danilov VI, Anisimov VM, Kurita N, Hovorun D (2005) MP2 and DFT studies of the DNA rare base pairs: the molecular mechanism of the spontaneous substitution mutations conditioned by tautomerism of bases. Chem Phys Lett 412:285–293
Al-Sehemi AG, El-Gogary TM, Wolschann KP, Koehler G (2013) Structure and stability of chemically modified DNA bases: quantum chemical calculations on 16 isomers of diphosphocytosine. ISRN Phys Chem 2013:1–11
Brovarets OO, Yurenko YP, Hovorun DM (2015) The significant role of the intermolecular CH···O/N hydrogen bonds in governing the biologically important pairs of the DNA and RNA modified bases: a comprehensive theoretical investigation. J Biomol Struct Dyn 33:1624–1652
Singh V, Fedeles BI, Essigmann JM (2015) Role of tautomerism in RNA biochemistry. RNA 21:1–13
Gardner N, Magers D, Hills G Jr (2014) Theoretical study of tautomeric and ionizing effects of guanine, cytosine, and their methyl derivatives. Struct Chem 25:347–353
Srivastava R (2019) The role of proton transfer on mutations. Front Chem 7:1–17
Farrel A, Murphy J, Guo J (2016) Structure-based prediction of transcription factor binding specificity using an integrative energy function. Bioinformatics 32:306–313
Mignon P, Loverix S, Steyaert J, Geerlings P (2005) Influence of the p–p interaction on the hydrogen bonding capacity of stacked DNA/RNA bases. Nucleic Acids Res 33:1779–1789
Jena NR, Bansal M, Mishra PC (2016) Conformational stabilities of iminoallantoin and its base pairs in DNA: implications for mutagenicity. Phys Chem Chem Phys 18:12774–12783
Jurecka P, Sponer J, Cerny J, Hobza P (2006) Benchmark database of accurate (MP2 and CCSD(T) complete basis set limit) interaction energies of small model complexes, DNA base pairs, and amino acid pairs. Phys Chem Chem Phys 8:1985–1993
Riley KE, Pitonak M, Cerny J, Hobza P (2010) On the structure and geometry of bimolecular binding motifs (hydrogen-bonding, stacking, X–H···π): WFT and DFT calculations. J Chem Theory Comput 6:66–80
Marek PH, Szatylowicz H, Krygowski TM (2019) Stacking of nucleic acid bases: optimization of the computational approach—the case of adenine dimmers. Struct Chem 30:351–359
Stauffer MT (2016) Applications of molecular spectroscopy to current research in the chemical and biological sciences. InTechopen, London
Greve C, Preketes NK, Fidder H, Costard R, Koeppe B, Heisler IA, Mukamel S, Temps F, Nibbering ETJ, Elsaesser T (2013) N–H stretching excitations in adenosine–thymidine base pairs in solution: base pair geometries, infrared line shapes, and ultrafast vibrational dynamics. J Phys Chem A 117:594–606
Madzharova F, Heiner Z, Gühlke M, Kneipp J (2016) Surface-enhanced hyper-Raman spectra of adenine, guanine, cytosine, thymine, and uracil. J PhysChem C 120:15415–15423
Shang Z, Ting DN, Wong YT, Tang YC (2007) A study of DFT and surface enhanced Raman scattering in silver colloids for thymine. J Mol Struct 826:64–67
Roberts GCK (2013) Infrared spectroscopy of DNA. Springer, Berlin
Heidari A (2016) An analytical and computational infrared spectroscopic review of vibrational modes in nucleic acids. J Anal Pharm Chem 3:1–14
Li C, Yang Y, Li D, Liu Y (2017) A theoretical study of the potential energy surfaces for the double proton transfer reaction of model DNA base pairs. Phys Chem Chem Phys 19:4802–4808
Tsolakidis A, Kaxiras E (2005) A TDDFT study of the optical response of DNA Bases, base pairs, and their tautomers in the gas phase. J Phys Chem A 109:2373–2380
Varsano D, Felice RD, Marques MAL, Rubio A (2006) A TDDFT study of the excited states of DNA bases and their assemblies. J Phys Chem B 110:7129–7138
Piqueras MC, Merchan M, Crespo R (2002) Conformational effects on the ultraviolet absorption spectrum of n-tetrasilane: multistate complete active space second-order perturbation theory treatment. J Phys Chem A 106:9868–9873
Xiaomei X, Xiaolei L, Hanhan L, Kelin L, Wei L (2016) Effect of conformation on UV–Vis absorption spectra of disazo reactive red dyes. Wuhan Univ J Nat Sci 21:1–6
Saha S, Quiney HM (2017) Solvent effects on the excited state characteristics of adenine–thymine base pairs. RSC Adv 7:33426–33440
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA et al. (2009) Gaussian 09, Revision A.02, Gaussian, Inc., Wallingford
Chakraborty S, Lima BC, Silva AM, Chaudhuri P (2017) Effect of hydrogen-bonded interactions on the energetics and spectral properties of the astromolecule aminoacetonitrile. Int J Quantum Chem 118:1–13
Dennington R, Keith T, MillamJ (2009) Gaussview, Version 5, Semichem Inc., K. S. Shawnee Mission
O’Boyle NM, Tenderholt AL, Langner KM (2008) GaussSum 3.0. J Comput Chem 29:839–845
Simon S, Duran M, Dannenberg JJ (1996) How does basis set superposition error change the potential surfaces for hydrogen-bonded dimers? J Chem Phys 105:11024–11031
Espinosa E, Molins E (2000) Retrieving interaction potentials from the topology of the electron density distribution: the case of hydrogen bonds. J Chem Phys 113:5686–5694
Biegler KF, Schonbohm J, Bayles D (2001) AIM2000: a program to analyze and visualize atoms in molecules. J Comput Chem 22:545–559
Lu T, Chen F (2012) Multiwfn: a multifunctional wave function analyzer. J Comput Chem 33:580–592
Wałęsa R, Kupka T, Broda MA (2015) Density functional theory (DFT) prediction of structural and spectroscopic parameters of cytosine using harmonic and anharmonic approximations. Struct Chem 26:1083–1093
Gu J, Leszczynski J (2000) Structures and properties of mixed DNA Bases Tetrads: nonempirical ab Initio HF and DFT studies. J Phys Chem A 104:1898–1904
Nosenko Y, Kunitski M, Stark T, Göbel M, Tarakeshwar P, Brutschy B (2013) Vibrational signatures of Watson–Crick base pairing in adenine–thymine mimics. Phys Chem Chem Phys 15:11520–11530
Sjoberg P, Politzer P (1990) Use of the electrostatic potential at the molecular surface to interpret and predict nucleophilic processes. J Phys Chem 94:3959–3961
Acknowledgements
S. Gop and R. Sutradhar would like to acknowledge the Department of Science and Technology (DST) for providing financial support in the form of DST INSPIRE Fellowship. (IF 170013, IF 160533). S. Chakraborty would like to acknowledge the financial assistance of SERB, DST (SERB Project No. SR/FTP/PS-073/2010).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gop, S., Sutradhar, R., Chakraborty, S. et al. Tautomeric effect of guanine on stability, spectroscopic and absorbance properties in cytosine–guanine base pairs: a DFT and TD-DFT perspective. Theor Chem Acc 139, 34 (2020). https://doi.org/10.1007/s00214-020-2551-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00214-020-2551-x