Abstract
PIWI-interacting RNAs (piRNAs) have received a lot of attention for their functions in cancer research. This class of short non-coding RNAs (ncRNA) has roles in genomic stability, chromatin remodeling, messenger RNA (mRNA) integrity, and genome structure. We summarized the mechanisms underlying the biogenesis and regulatory molecular functions of piRNAs. Among all piRNAs studied in cancer, this review offers a comprehensive analysis of the emerging roles of piR-823 in various types of cancer, including colorectal, gastric, liver, breast, and renal cancers, as well as multiple myeloma. piR-823 has emerged as a crucial modulator of various cancer hallmarks through regulating multiple pathways. In the current review, we analyzed several databases and conducted an extensive literature search to explore the influence of piR-823 in carcinogenesis in addition to describing the potential application of piR-823 as prognostic and diagnostic markers as well as the therapeutic potential toward ncRNA precision.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
PIWI-interacting RNAs (piRNAs) are a newly discovered class of short non-coding RNAs (ncRNAs) in germ and somatic cells that consist of 26–30 nucleotides (nt) with a uridine base at the 5′-terminal or an adenosine base at the tenth position and a 2′-O-methyl at the 3′-terminal, that seem to have distinctive properties of all piRNAs (Aravin et al. 2006). These short chains of RNA were identified in germline cells and recognized as important modulators of germline maintenance (Huang et al. 2013a). Currently, piRNAs have a crucial role in controlling genomic expression in various disorders, including cancer, via several mechanisms such as transposon silencing, epigenetic modifications, and chromatin remodeling (Jia et al. 2022). The family of Argonaute proteins, divided into the Ago and PIWI subfamilies, is essential to short RNA-mediated gene regulatory pathways. Argonaute 1 (Ago1) and 2 (Ago2) members of the Ago subfamily of Argonaute proteins are known to be involved in small interfering RNA (siRNA)-mediated messenger RNA (mRNA) degradation and microRNA (miRNA)-mediated gene regulation, respectively (Yang et al. 2007). In Drosophila, the subfamily of PIWI proteins consists of Aubergine (Aub), Piwi, and Ago3, which are involved in retrotransposon silencing as well as germline stem cells’ maintenance and self-renewal. Ago3 and Aub are expressed in the cytosolic portion of germ cells, but Piwi is the only one of the three PIWI proteins, which is predominantly nuclear in germline and somatic cells present in the gonad and regulates piRNA synthesis in both types of cells (Huang et al. 2014a). Four PIWI proteins are expressed in humans, which are PIWI-like protein 1–4 (PIWIL1-4) (Tan et al. 2015). PIWI proteins influence transposon silencing, genome rearrangement, and epigenetics by creating complexes with piRNAs that control methylation and subsequently transposable elements (TEs) inhibition. Several types of cancer are either inhibited or promoted by transposon silencing and many other associated phenomena, including epigenetic modifications (Cheng et al. 2019). Although significant progress has been made in cancer research concerning diagnostics and therapeutics, the complete remission and survival of cancer patients remains a significant challenge. There is mounting evidence connecting piRNA/PIWI to cancer prognosis and carcinogenesis (Liu et al. 2019).
Search strategy
A manual online search into two medical e-databases PUBMED and google scholar for (“piR-823” OR “piRNA-823”) AND (“piR-823 in Cancer”) AND (“piRNA in Cancer”) AND (“in silico”) AND (“Cancer hallmarks”) AND (“SNPs”) AND (“Piwi proteins”) AND (“Prognostic marker”) AND (“Diagnostic marker”) AND (“Nanoparticles”) AND (“Exosomes”) was done on June, 2023. Priority was given to meta-analysis, randomized clinical studies, systematic review, original papers, and narrative reviews, since, but not limited to, 2010.
piRNA biogenesis
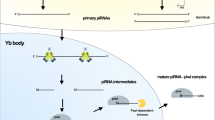
The way that piRNAs differ from miRNAs is that the piRNA precursors are often single-stranded and created from particular genome sites that include repeated motifs, and their coordinated signaling pathway is not dependent on dicer (Huang et al. 2014b). According to their origin, piRNAs are grouped into three types: (1) repeat-associated piRNAs, also known as “transposon-derived piRNAs,” from intergenic loci known as piRNA clusters and are rich in transposon segments, (2) mRNA-derived piRNAs produced from the 3′ UTR of mRNAs, and (3) long non-coding RNA (lncRNA)-derived piRNAs (Han and Zamore 2014). Transposon-derived piRNAs have a considerably better understanding of their biogenesis and functions than the other two types (Moyano and Stefani 2015). Following transcription, primary synthesis and “ping-pong” amplification are the main processes that transform piRNA primary transcripts into mature piRNAs (Jia et al. 2022).
The primary synthesis is based on the RNA polymerase II-mediated transcription of short sequences from piRNA precursors known as clusters. These transcripts are transported to the cytoplasm, where zucchini (Zuc) and its cofactor protein minotaur (Mino) trim them into smaller sequences to generate piRNA intermediates (Jia et al. 2022). The PIWI protein then binds to piRNA intermediates to form the PIWI/piRNA complex, which returns to the nucleus to initiate transcription-silencing mechanisms to inhibit the expression of its target genes by pairing the complementary bases of piRNAs and DNA (Czech and Hannon 2016).
The “ping-pong” amplification process produces a huge number of piRNAs in the cytoplasm. Through this process, piRNAs generate Ago3/piRNA or Aub/piRNA complexes by binding to Ago3 or Aub proteins instead of PIWI proteins (Xiol et al. 2014). The sequences in these complexes complement each other. In this way, an Ago3/piRNA complex identifies and trims an RNA sequence, producing a new RNA sequence that serves as a substrate for the formation of a new piRNA able to bind an Aub protein. Similarly, the resulting Aub/piRNA complex cuts a complementary RNA sequence, generating more RNA molecules, which in turn produce new Ago3/pirn complexes (Jia et al. 2022). As a result, the piRNA cytoplasmic product acts as a substrate for another piRNA molecule in an amplification manner. Finally, these piRNAs bind to PIWI proteins and return to the nucleus, where they inhibit the transcription of the targeted genes (Assumpção et al. 2015).
piRNA regulatory functions
It has become evident that piRNAs are considered transcriptional regulation molecules through transposon silencing in addition to gene and protein regulation (Fig. 1).
Transcriptional gene silencing (TGS)
The piRNA/PIWI complex exploits histone modifications and DNA methylation to control to transcriptional gene silencing (TGS). The piRNA/PIWI complex, after being produced in the cytoplasm, moves into the nucleus to combine with the genomic target to recruit silencing molecules and trigger the beginning of the TGS. DNA methyl transferase (DNMT) is recruited by the piRNA/PIWI complex to methylate sites containing CpG and alter the transcription function (Kuramochi-Miyagawa et al. 2008). Likewise, piR-30473 is implicated in the carcinogenesis of diffuse large B-cell lymphoma by modulating methylation of m6A RNA (Han et al. 2021). In primary multiple myeloma (MM) cells, de novo DNMT3A and DNMT3B were directly recruited by piR-823 to initiate overall DNA methylation (Yan et al. 2015).
Posttranscriptional gene silencing (PTGS)
piRNAs are currently known to engage in post-transcriptional functions resembling miRNAs by acting on other cytoplasmic RNAs such as mRNA, transcribed pseudogenes, and lncRNA (Liu et al. 2019). For example, piR-55490 could bind to the mammalian target of rapamycin (mTOR), causing mRNA damage and thereby preventing lung cancer incidence and progression (Peng et al. 2016). Also, piR-1245 promotes tumor progression by binding to a class of downstream target genes that control cell survival, resulting in RNA silencing via aberrant base-pairing (Weng et al. 2018). Likewise, piR-823 binds to eukaryotic initiation factor 3 B (EIF3B) and upregulates the expression of transforming growth factor-1beta (TGF-1β) to initiate hepatic stellate cells during hepatic fibrosis (Tang et al. 2018).
Posttranslational modifications (PTMs)
Post-translational modifications occur in the cytoplasm, where the piRNA/PIWI complex interacts with various transcription factors and controls their post-translational alterations, such as phosphorylation. In colorectal cancer (CRC), piR-823 promotes tumorigenesis by increasing the activation and transcriptional function of heat shock factor 1 (HSF1) (Yin et al. 2017). Moreover, the piR-54265/PIWIL2 complex interacts with the signal transducer and activator of transcription 3 (STAT3) to produce a complex that activates the phosphorylation of STAT3, thereby promoting CRC cell proliferation, metastasis, and chemoresistance (Mai et al. 2018).
Transposone silencing
Transposons are mobile DNA components that account for less than 2% of the individual genome and utilize the genome and replicative machinery of the host cells for their existence and proliferation (Yuan et al. 2021). TEs can introduce new genetic material into the genome and frequently induce DNA modifications such as deletion, duplication, or inversion (Cheng et al. 2019). piRNAs preserve genomic integrity and stability through transposon silencing by forming an RNA-induced silencing complex (RISC). piRNAs bind to a certain transposon sequence through base complementarity. This mechanism effectively inhibits genomic transposition elements, preventing the cellular genes from being destroyed (Yuan et al. 2021). Furthermore, RISC can bind to PIWI proteins and target transposon transcripts in order to silence them and utilize active transposon transcripts as precursors for piRNA amplification through the ping-pong mechanism (Moyano and Stefani 2015). Surprisingly, new research has revealed that transposons, via the PIWI-piRNA pathway, actively control mRNAs and lncRNAs expression. Furthermore, pseudogenes control the expression of their related mRNAs via the piRNA/PIWI pathway (Wang and Lin 2021). According to a previous study, transposon regions in lncRNAs act as the targets for piRNAs formed from transposon-derived lncRNAs, which mark these lncRNAs for decay. A global analysis of the transposon dispersion in the human genome revealed that 83.4% of lncRNAs overlap with the minimum of one transposon, compared to only 6.2% of protein-coding sequences (Kelley and Rinn 2012). Numerous organisms have shown piRNAs to be involved in the transposon control of lncRNAs. The overexpression of transposon-containing lncRNAs in Miwi mutant mouse spermatocytes suggests that the piRNA pathway is the mechanism by which transposon regulates lncRNA expression (Vourekas et al. 2016).
piRNA datasets and target predictions
piRNA–mRNA and piRNA–lncRNA interactions
The piRNA-mediated cleavage of TEs is a well-known function of piRNA. Nevertheless, recent studies have revealed the involvement of piRNAs in controlling the expression of mRNAs and lncRNAs. The cleavage of piRNA-targeted mRNA occurs at the tenth position from the 5′ terminal of the targeted piRNAs, which are antisense to the mRNA sequence (Wang and Lin 2021). piRNAs may be able to silence an mRNA by targeting its protein-coding sequence (CDS). For instance, Aub directly and piRNA-dependently targets the 3′ UTR and CDS of maternal mRNAs in the Drosophila to promote mRNA degradation (Barckmann et al. 2015). The incorporation of transposon sequences into mRNAs as piRNA targets involves dynamic mechanisms regulating mRNA expression, at least for certain cell types (Watanabe et al. 2015). piRNA-mRNAs interactions involve canonical mRNA decay machinery. Therefore, piRNA-mediated mRNA degradation also involves contemporaneous deadenylation and decapping of mRNAs in addition to PIWI slicing of the target mRNAs (Wang and Lin 2021). The PIWI/piRNA complex trims targeted lncRNAs like how it slices mRNAs. Then, mature piRNAs are integrated into PIWI proteins, which enable the PIWI complex to destroy target lncRNAs by targeting those lncRNAs with complemented transposon sequences (Sytnikova et al. 2014). piRNAQuest (http://dibresources.jcbose.ac.in/zhumur/pirnaquest2/start.php), a comprehensive database that provides information on piRNA features and piRNA expression in relation to various tissues. It offers a diverse narrative focusing on pseudogenes and predicts piRNA–mRNA and piRNA–lncRNA interactions and overlaps in various diseases and cancer. For instance, it showed target pairing between piR-823 and lncRNA for the BRAC1 gene in breast cancer (BC), SH3BP2 in renal cell carcinoma (RCC), and ANKRD28 in hepatocellular carcinoma (HCC) (Fig. 2).
pirnaPre (http://www.regulatoryrna.org/software/piRNA/piRNA_target_mRNA/index.php) and pirScan (http://cosbi4.ee.ncku.edu.tw/pirScan/), tools designed for identifying and predicting piRNA targets. IsopiRBank (http://mcg.ustc.edu.cn/bsc/isopir/index.html) is a comprehensive online tool that offers piRNA isoform analysis, enrichment analysis, and target prediction. Moreover, piRNA targets and functions can be explained by the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of piRNA targets (Zuo et al. 2019). piRBase (http://bigdata.ibp.ac.cn/piRBase/) is another prediction tool for piRNA targets in lncRNAs in mice. There are several tools available for predicting miRNA targets; however, there are not many for piRNA targets. Research on prediction algorithms and tools for piRNAs and their targets is still in the early stages. Several tools, databases, and algorithms are dedicated to the detection and analysis of piRNA as collected in Table 1. Although piRNA bioinformatics research has evolved, further studies are required to overcome the limitations of the computational tools of piRNA (Liu et al. 2021). This is accomplished first by strengthening the development of piRNA databases since different databases have different names for piRNAs, making piRNA searches more difficult. Specifically, piR-823, one of the most studied piRNAs, has different aliases (piR-31143, piR-1282, piR-1119, piR-000823, piR-30928) stored in different databases such NCBI, piRNABank, piRBase, piRNAQuest, and piRNAdb. Secondly, since many databases, including pirScan and piRBase, lack data on human species, computational biologists should investigate piRNA to increase the diversity of species studied. Finally, piRNA-target prediction and disease association data should be improved to facilitate computational and experimental research.
In silico identification and expression of piR-823
Abnormal expression of piR-823 has been associated with several tumors and may have an oncogenic or tumor suppressor effect on the onset, progression, and spread of cancer. From the piRNAdb database (https://www.pirnadb.org/index) and UCSC Genome Browser (https://genome.ucsc.edu/index.html), piR-823 contains 32 bases located in chromosome 19 (Fig. 3). Using piRNAQuest V2., piR-823 is highly expressed in different tissues as shown in Fig. 4a and in different cancers as shown in Fig. 4b. Moreover, PiRpheno V2.0 database (http://www.biomedical-web.com/pirpheno/pirpheno.html) showed the expression profile of piR-823 in various diseases and cancer as shown in Fig. 5. piR-823 was found to be associated either positively or negatively with 92 different diseases and cancers. Hematopoietic, lymphoid, and myeloma neoplasm were among the highest confidence score correlation. Interestingly, piR-823 was found to be associated with neurodegenerative diseases such as Alzheimer’s disease.
Illustration of genomic localization of piR-823 using piRNAdb database (https://www.pirnadb.org/information/pirna/hsa-piR-1119) (Accessed May,2024)
Expression level of piR-823 (piRNA-30928) in a different tissues. The expression graph of piR-823 is represented in count per million (CPM). As shown, the highest expression of piR-823 was found in pancreatic tissues using piRNAQuest V.2 database http://dibresources.jcbose.ac.in/zhumur/pirnaquest2/expressn_view.php?organism=hsa&pirna_id=hsa_piRNA_30928&submit=Submit. b Different cancers. The expression graph of piR-823 is represented in count per million (CPM). As shown, the highest expression of piR-823 was found in breast cancer using piRNAQuest V2 http://dibresources.jcbose.ac.in/zhumur/pirnaquest2/expressn_view.php?organism=cancer&pirna_id=hsa_piRNA_30928&submit=Submit (Accessed May, 2024)
piR-823 expression in different human cancer using PiRpheno V2.0 (http://www.biomedical-web.com/pirpheno/Networklyzer.jsp) (Accessed May, 2024). The lines that are thicker signify a higher confidence level (ranging from 0 to 1) for the piRNA-disease relationship; whereas, the green lines show a negative correlation between the piRNAs and the illness phenotypes. The red lines show a positive correlation between the two. The piRNA-disease relationships with contradicting evidence are shown by the dashed lines
piR-823 interactions in different cancer types
Though the potential role of piR-823 in cancer is being highly investigated, its functional role in human cancer is poorly known. The expression and function of piR-823 in various human cancers have been summarized, as shown in Tables 2 and 3.
Colorectal cancer
As the most common form of gastrointestinal (GI) cancer, colorectal cancer (CRC) is now the third most common cause of cancer-related death. Owing to the diverse nature of CRC, several pathways, such as chromosomal instability, microsatellite instability, epigenetic alterations (Abd El Fattah et al. 2023; El-Sheikh et al. 2023; Emam et al. 2022; Rizk et al. 2024), and abnormal expression of tumor oncogenes or suppressor genes, can influence CRC initiation and proliferation (Ameli Mojarad et al. 2022). However, the majority of patients are diagnosed in late stages due to insufficient prognostic and diagnostic biomarkers, reducing their chances of receiving early treatment; thus, identifying robust and precise therapeutic indicators is essential for managing CRC (Mojarad and Mojarad 2021). piRNAs, through their binding to PIWI, have a vital role in the pathogenesis of CRC, and genetic variants in piRNAs may modulate CRC susceptibility (Henaoui et al. 2017). Cancer research emphasizes finding novel, reliable diagnostics for the early detection of GI malignancies and therapeutic targets that function as targeted treatments to lower mortality. Consequently, studies have demonstrated that piRNAs have important clinical implications as CRC therapeutic targets and tools for diagnosis. Thirty-one deregulated piRNAs, including piR-5937 and piR-28876, showed adequate diagnostic performance in large-scale piRNA expression analysis in serum samples from colon cancer patients compared to normal groups, implying the clinical utility of piRNAs as early diagnostic markers for differentiating between cancer and normal individuals (Vychytilova-Faltejskova et al. 2018). piR-823 is one of the piRNAs that contributed to the tumorigenesis of CRC and could be utilized as a possible CRC therapeutic target. Upregulation of piR-823 promotes both proliferation and tumorigenesis in CRC tissues by upregulating the expression of HSF1, a potent carcinogenesis trigger, and by influencing the phosphorylation of STAT3 (Yin et al. 2017). In a different study, it was found that piR-823 targeted PTEN-induced kinase 1 (PINK1), promoting the proteasome-mediated degradation of PINK1 and preventing mitophagy and cell death in CRC cell lines. piR-823 reduced the ubiquitination of the mitochondrial proteins mitofusin (MFN2) and translocator of the outer mitochondrial membrane (TOM20), which may have contributed to inhibiting mitophagy mediated by PINK1/Parkin signaling (Wang et al. 2022b). Similarly, piR-823 increases glucose-6-phosphate dehydrogenase (G6PD) expression, which in turn increases the consumption of glucose by cancer cells while decreasing the amount of intracellular reactive oxygen species (ROS). This suppresses the ubiquitination of hypoxia-inducible factor 1 alpha (HIF-1 α). Therefore, via modifying the G6PD/HIF-1α pathway, piR-823 increases CRC cell growth, invasion, and resistance to apoptosis (Feng et al. 2020). Because of the overexpression of piR-823 in CRC patients’ serum and tissues, it holds promise as a convenient, non-invasive screening technique for CRC (Sabbah et al. 2021). piR-823 can be utilized for clinical diagnosis of CRC in addition to carcinoembryonic antigen (CEA) (Tong et al. 2023).
Hepatocellular carcinoma
Hepatocellular carcinoma (HCC) is the second most common cause of cancer-related deaths in both men and women. It develops as a consequence of a progressive process that frequently starts with the morphologically identifiable low- and high-grade dysplastic nodules within a cirrhotic liver developing premalignant lesions. Numerous studies have identified and characterized many abnormal gene expressions, single-nucleotide polymorphisms (SNPs), and noncoding RNAs (ncRNAs) in HCC (Abaza et al. 2023; El-Aziz et al. 2023; El-Derany et al. 2016; Hammad et al. 2023). The multifactorial characteristics of the pathways, which emphasize the importance of epigenetic variables including miRs, lncRNAs, and piRNAs, may help to understand the pathogenesis of HCC (Eldosoky et al. 2023). piR-823 is a novel player in the molecular processes that lead to hepatocarcinogenesis. Whereas, piR-823 levels increase steadily from cirrhosis to low-grade and then high-grade dysplastic nodules and HCC, suggesting that they could be new helpful biomarkers for differentiating dysplastic and neoplastic liver lesions (Rizzo et al. 2016). Moreover, piR-823 may influence liver fibrosis by activating hepatic stellate cells (HSCs). Overexpression of piR-823 increased HSCs viability and proliferation by increasing the generation of alpha-smooth muscle actin (α-SMA) and collagen type 1 alpha 1 (COL-1a1). piR-823 interacts with EIF3B, and this complex induces TGF-1β expression, activating dormant HSCs. Thus, inhibiting piR-823 and HSCs might represent a new strategy for treating liver fibrosis (Tang et al. 2018). Similarly, piR-823 might act as a therapeutic target in HCC. As a recent study revealed that 4-hydroxycoumarin treatment caused upregulation of piR-823, matrix metallopeptidases (MMP-2, and MMP-9) expression, which are important features of cellular survival mechanisms that led to the loss of viability, adhesion, and proliferation of HepG2 cells (Öner et al. 2022). Furthermore, 1,25-dihydroxy vitamin D therapy increased proliferation, adhesion, differentiation, and telomerase activity in HepG2 cells through increasing piR-823 expression while decreasing HIF-1α, MMP-2, and MMP-9 expression. Unfortunately, the active form of vitamin D may influence the epigenetic mechanisms of HCC cells via piR-823 expression and may not be beneficial for as an anti-cancer in HCC patients (oner 2021).
Gastric cancer
With over one million new cases identified each year, gastric cancer (GC) is the fifth common cause of cancer-related deaths worldwide (Ameli Mojarad et al. 2022). The overall survival of people with GC has improved as early detection of cancer has increased, and radical surgery has become more widely used. However, the prognosis for advanced GC is still poor, and choices for safe and effective adjuvant therapy are limited (Li 2014). As a result, finding valuable early markers is critical for GC diagnosis and prognosis. The abnormal expression of piRNAs and PIWI proteins has been reported in GC tissues, implying their crucial role as a promising biomarker in gastric tumorigenesis. piR-823 expression was considerably lower in GC patients’ peripheral blood compared to healthy controls. With a correlation found between the levels of piR-823 and both distant metastasis and tumor node metastasis (TNM) indicating that piR-823 detection may serve as a prognostic marker for GC (Cui et al. 2011). Overexpression of piR-823 inhibited the growth of both GC cells and transplanted tumors, suggesting the involvement of piR-823 in the development of GC and implying that piR-823 may have a tumor suppressor effect on the GC progression (Cheng et al. 2012). Both piR-823 and piR-651 were more sensitive for GC than CEA and carbohydrate antigen 19–9 (CA19-9) (Cui et al. 2011). piRNAs are not easily degraded because they are short fragments, and the levels of piR-651 and piR-823 in blood samples are reasonably stable and may be detected and isolated from body fluids as they can cross easily through the cell membrane (Chalbatani et al. 2019). PIWIL1 expression has elevated in GC cells, indicating that a high level of PIWIL1 expression may be a crucial prognostic indicator for predicting the survival of GC patients. By modulating the expression of oncogenes and tumor suppressor genes, PIWIL1 may accelerate GC pathogenesis and progression (Guo et al. 2020). The precise mechanisms by which PIWIL1 and piR-823 contribute to the proliferation and metastasis of GC are still unclear. Nonetheless, favorable prospects are indicated by the aberrant expression of piRNA and PIWI proteins in GC tissues compared to normal tissues.
Breast cancer
Breast cancer (BC) is one of the most common malignancies and the leading cause of cancer-related mortality in women worldwide. According to the expression analysis profiles of progesterone receptor (PR), estrogen receptor (ER), and human epidermal growth factor receptor 2 (HER2), BC is divided into four subtypes: luminal, HER positive, basal-like, and normal-like (Ding et al. 2021). Despite the significant improvements in human BC therapeutics, tumor recurrence and metastasis remain incurable, owing to the chemo-resistance from the ncRNA effect (Mahmoud et al. 2021b), as well as more tumor growth promotion from activated oncogenes vs tumor suppressor genes inhibition (Mahmoud et al. 2021a; Rezaeean et al. 2021; Salama et al. 2021). All these effects are regulated via ncRNAs and one of which to be explored and examined is piRNAs. According to a recent study, by methylating a particular gene, piRNAs can influence the proliferation, metastasis, cancer development, and prognosis of BC. This suggests that piRNAs may be a promising target for therapy and a novel class of biomarkers for the early detection and prognosis of BC (Öner et al. 2016). piR-823 is a novel modulator of cancer stem cells (CSCs) in luminal BC since suppression of piR-823 significantly reduced the growth of tumors and the proliferation of cancer cells both in vitro and in vivo by controlling DNMTs expression and by increasing the DNA methylation levels of tumor suppressor genes like adenomatous polyposis coli (APC), consequently, activating the WNT signaling pathway, which in turn induces cancer stemness in BC (Ding et al. 2021). piR-823 reacted to the hormonal therapy in a cancer subtype-specific way, as external treatment of estrogen increased the expression of piR-823 in the basal-like subtype triple-negative (ER, PR, and HER2 negative) (Öner et al. 2016). Meanwhile, decreasing the expression of piR-823 levels in the luminal subtype (ER and PR). As a result, high piR-823 expression promotes tumorigenesis in luminal BC, and piR-823 suppression may be used to predict patients’ therapeutic response to hormone therapy. A recent study revealed that inhibition of piR-823 increased ER-α expression while decreasing the expression of human telomerase reverse transcriptase (hTERT) and the phosphatidylinositol 3-kinase (PI3K)/AKT/mTOR pathway, which influences cell proliferation and cellular properties in triple-negative BC cells (oner 2022).
Multiple myeloma
Multiple myeloma (MM) is a B-cell-derived cancer that causes clonal proliferation of plasma cells in the bone marrow (Khalili et al. 2023; Robak et al. 2018). Malignant plasma cell proliferation and MM development are associated with aberrations in the bone marrow microenvironment. Secondary leukemia and extramedullary disease might result from the separation of MM cells from the bone marrow microenvironment (Furukawa and Kikuchi 2015). CSCs persisting in the bone marrow of MM patients are one of the underlying mechanisms of drug resistance and relapse because these cells have an infinite potential for self-regeneration and drug resistance (Li et al. 2021). Another mechanism is genetic and epigenetic abnormalities, including DNA methylation and histone modifications, in various genes, particularly tumor suppressors (Robak et al. 2018). CSCs and myeloid-derived suppressor cells (MDSCs), major cellular factors in the tumor microenvironment in MM, can alter the tumor phenotype and influence patient prognosis and outcome. Granulocytic-MDSCs (G-MDSCs), a type of MDSC, increased the expression of piR-823, which in turn enhanced DNA methylation and promoted the tumorigenesis of MM cells (Ai et al. 2019). The ability of G-MDSCs to retain stemness diminished when silencing piR-823 in MM cells, which reduced tumor growth and angiogenesis in vivo, implying that piR-823 might be a unique anti-cancer therapy that targets both G-MDSCs and CSCs in the MM microenvironment (Ai et al. 2019). Endothelial cells (ECs) in the tumor microenvironment are highly required for the proliferation, survival, and dissemination of MM cells (Korn and Méndez-Ferrer 2017). Extracellular vehicles (EVs) derived from MM consist of functional proteins, mRNAs, miRNAs, and piRNAs (Raposo and Stoorvogel 2013). PiR-823 levels were significantly greater in peripheral EVs obtained from patients with MM (Li et al. 2019), suggesting a potential role for piR-823 in the diagnosis and prognosis of MM. piR-823 delivered to ECs by EVs derived from MM stimulates EC spread, angiogenesis, and proliferation which may eventually contribute to the progression of MM (Li et al. 2019). Another study reported that silencing piR-823 in MM cells reduced the release of vascular endothelial growth factor (VEGF), resulting in diminished angiogenic activity (Yan et al. 2015). All these results support the oncogenic involvement of piR-823 in the biology of MM and justify the need for exploring piRNA-targeted therapeutic approaches in MM.
Genitourinary cancers
Studies on renal cancer have revealed a strong correlation between piRNA and the disease’s prognosis and metastasis. Additionally, in RCC, the most common kidney cancer in adults, the expression levels of PIWIL1, PIWIL2, and PIWIL4 gradually decrease with increasing clinical stage and a worsening prognosis (ILIEV et al. 2016). piR-823 has a complex function in the tumorigenesis and prognosis of RCC. Preliminary results show that piR-823 is highly expressed in urine and has a great diagnostic value in RCC patients (ILIEV et al. 2016). Another study reported the association of PIWIL1 in the piR-823-mediated pathogenesis of RCC through the deregulation of PIWIL1 and piR-823 in RCC tumor tissue with a positive correlation, as well as a substantial association has been observed between PIWIL1 and PIWIL2 and PIWIL4 expression and RCC survival (Slaby et al. 2016). Gender-specific steroid hormones are important in the development and pathophysiology of cancers of the reproductive system, such as androgen-dependent or androgen-independent prostate cancer in men (Feldman and Feldman 2001). piR-823 is one of the piRNAs important for cancers that are gender-dependent. Its overexpression in cancer is thought to be mediated by hormones (Öner et al. 2016). Increasing hormone levels, such as androgen, in prostate cancer cells, showed that the elevated piR-823 expression exhibited oncogenic properties in both androgen-dependent and androgen-independent prostate cancer cell lines and that this property increased as hormone levels increased (Öner et al. 2016).
piR-823 SNPs in cancer
While most of the research has been on the aberrant epigenetics and gene expression of piRNAs in cancer, a few studies have also documented the relation between piRNAs and SNPs along with other genetic variations. Finding SNPs in particular or PIWI proteins was linked to a patient’s risk of developing cancer (Yao et al. 2022). Seven SNPs in nine piRNAs were evaluated for CRC susceptibility, and SNP rs11776042 in piR-015551 was found to be substantially linked to a lower risk of CRC. Results suggested that the rs11776042 thymine to cytosine (T > C) substitution affected the energy of the piRNA secondary structure and that might influence the role of piRNA on CRC development (Chu et al. 2015). Genotypic analysis of several SNP-containing piRNAs indicated a strong association between a higher risk for developing BC and SNP rs1326306 G > T in piR-021285 (Fu et al. 2015). A post-genome-wide association study (GWAS) and functional analyses of piRNAs in glioma-genesis revealed that SNP rs147061479 in piR-598 enhances the incidence of gliomas by eliminating piR-598 tumor-suppressive function (Jacobs et al. 2016). Another substantial evidence supporting the potential role of piRNA variants in lung cancer was found in a different post-GWAS study that identified SNP (rs1169347) as strongly related to lung cancer risk, and it can be matched into piR-5247 and piR-5671 (Ye 2016). Compared to piRNAs, SNPs in PIWI and piRNA-PIWI pathway-related genes are more prevalent and significantly linked to cancer risk (Lin et al. 2019; Roy et al. 2020; Zhang et al. 2016). SNPs identified specifically in piR-823, however, have not yet been reported, which calls for more thorough molecular research and genotyping screening. Exploring the association between ncRNAs-SNPS, and more precisely, piR-823 SNPs, and the cancer mechanism(s), is important to investigate to link certain polymorphisms to changes in cancer incidence, progression, or remission in the future. This represents a step toward the precision of ncRNA.Footnote 1
Additionally, because piRNA variants have been experimentally verified as a novel class of biomarkers and therapeutic targets, it is now crucial to identify and understand the connections between piRNA variants and human diseases. Numerous computational databases and techniques have been developed to annotate piRNA variants due to their clinical and functional significance. For example, ncRNAVar (http://www.liwzlab.cn/ncrnavar/) offers association data between human diseases and validated ncRNA variants using computational annotation and manual curation of 2650 publications (Zhang et al. 2021). Also, piRNA-eQTL (http://njmu-edu.cn:3838/piRNA-eQTL/) provides analysis between SNPs and piRNA expression over 33 types of cancer using the cancer genome atlas (TCGA) program (Xin et al. 2021). Researchers may find it useful to use piRSNP (http://www.ibiomedical.net/piRSNP/) to thoroughly characterize SNPs associated with piRNA in humans and mice, as this information can be used to study piRNA functions in the future (Liu et al. 2023). These databases are a valuable tool for motivating researchers to investigate more ncRNA variants, which helps in understanding the roles that piRNAs and genetic variants play in cancer development.
piR-823 clinical utility in cancer
piR-823 as non-invasive cancer bio-molecular marker
piRNAs have received a lot of attention for their potential to be utilized in the diagnosis and prognosis of various types of cancers (oner 2022; Yan et al. 2015). Quantitative variations in the expression level of these piRNAs might help identify the origin of cancer tissue as a portion of piRNAs might be expressed in a way that is very cancer-specific (Cheng et al. 2011). A growing body of evidence revealed that piRNAs may be useful diagnostic and prognostic biomarkers in cancer, even though the proposed mechanisms need more investigation and may not fully explain piRNAs’ role as cancer biomarkers. piRNA expression in the body’s fluids, including blood, plasma, serum, saliva, sputum, and urine, is comparatively constant and readily detectable (Mei et al. 2013). This stability could be attributed to piRNAs’ 2′-O-methyl modification, which shields them from 3′ uridylation and truncation, two processes that affect the integrity of short RNAs (Jeong et al. 2021). Furthermore, piRNAs are just 26–32 nt long, a tiny RNA fragment that makes them more resistant to degradation than other large RNAs and allows them to easily cross various cell membranes. These properties indicate that piRNAs can be readily found and isolated from patient specimens (Chalbatani et al. 2019). As explained above, piR-823 overexpression in various cancers including colon, liver, renal, and breast cancer tissues compared to normal tissues, indicating that piR-823 may serve as a promising diagnostic biomarker for these cancers (Table 2). The prognosis of cancer has also demonstrated significant promise for piRNAs. Multiple studies revealed a correlation of piR-823 with TNM, distant metastasis, histopathology type, or cancer progression in several cancer types, implying that piR-823 could be an independent prognostic marker (Yan et al. 2015; Ai et al. 2019; Su et al. 2020; ILIEV et al. 2016). Another reason for utilizing piRNA as a biomarker is the encapsulation of certain piRNAs in the blood into exosomes and microvesicles, two types of extracellular vesicles, thereby protecting them from ribonuclease digestion. EV-piRNAs have shown promise as a diagnostic tool for cancer types (Goh et al. 2022). As identified, piR-823 aggregates in extracellular vesicles (EVs) from the peripheral blood of MM patients and cell lines. It is essential for the cellular interaction between the endothelial cells and myeloma cells in the tumor microenvironment. It is possible to successfully transfer EVs harboring piR-823 from MM cells to ECs and change their biological properties to establish an environment supportive of MM cells’ survival (Li et al. 2019). Collectively, aberrantly expressed piRNAs in human tumors represent a novel class of biomarkers for prognosis and diagnosis. Exosomes are one of the EVs produced from the multi-vesicular endosome pathway and are packed with biologically significant molecules like DNA, mRNA, and ncRNAs (Becker et al. 2016). Therefore, exosomes with a diameter ranging from 30 to 150 nm can serve as the best cargo for carrying information from cancer cells, and that information could be easily found in serum exosomes (Gu et al. 2020). Exosomes have gained researchers’ interest in cancer therapy because of their distinctive properties, including biocompatibility, minimal toxicity, circulatory stability, tumor specificity, and high tumor penetration capacity (Rizk et al. 2024). RNAs in plasma exosomes are differentially expressed and considered a promising biomarker for various cancer types; however, more research is needed to explore the novel content of exosomes especially piRNAs which account for 20–30% of total RNA (Ma et al. 2020). That suggests the great potential of exosomal piRNAs as an early non-invasive diagnostic and prognostic tool in cancer patients. In addition to CEA, serum exosomal piR-823 was discovered to be a reliable biomarker of CRC (Tong et al. 2023). Expression levels of piR-004800 in exosomes derived from bone marrow supernatant in MM patients and cells were associated with disease staging, which suggests that piR-004800 may be a marker for MM disease progression (Ma et al. 2020). Exosomal piR-25783 stimulates the TGF-β/SMAD2/3 pathway and encourages the secretion of different cytokines that play a significant role in the premetastatic microenvironment in metastatic ovarian carcinoma (Li et al. 2023). A total of 253 differentially expressed piRNAs were identified by small RNA sequencing analysis of piRNAs obtained from serum exosomes of HCC patients. These piRNAs were effective in differentiating HCC patients from non-tumor donors, suggesting that exosomal piRNAs could be potential diagnostic biomarkers for HCC (Rui et al. 2023). Exosomal piR-17560 produced by senescent neutrophils promotes the production of obesity-related protein in BC cells and provides tumor cells chemoresistance, suggesting that senescent neutrophils could be a viable target for therapy in the recurrence of BC (Ou et al. 2022). Exosomal piR-1089 is considered a quick, early, and easy noninvasive tool for neuroblastoma assessing metastasis. piR-1089’s mechanism in neuroblastoma cell proliferation and migration could provide more reliable experimental evidence for precision medicine (Wang et al. 2023). Exosomal piR-164586, piR-26925, and piR-5444 were significantly overexpressed in the serum of lung adenocarcinoma patients compared to healthy controls (Li et al. 2022). Recently, exosomes have been studied as potential nanocarrier therapies for a variety of diseases, including neurological conditions like Parkinson’s and Alzheimer’s. Exosomes’ small dimension allows them to pass through the BBB and into the CNS through a systemic mode of delivery. Exosomes allow Alzheimer’s disease cells to discard non-functional proteins, acting as “waste bags” in the process. Additionally, their RNA cargo can support the development of neural cells where piRNA and more precisely piR-823 would have potential worth testing.Footnote 2 In this promising area, more studies are required to clarify the association between piRNAs and cancers.
Therapeutic insights into piRNA
Several studies investigated the therapeutic applications of piRNAs in addition to their function as biomarkers. Despite the limited knowledge of the molecular pathways underlying piRNA activity in carcinogenesis, piRNA functional characteristics would make them excellent candidates for therapeutic purposes (Chen et al. 2021). To silence PIWI genes and eliminate negative consequences, specific piRNAs could be designed to bind PIWI proteins at the pre-transcriptional level. The PIWI/piRNA pathway may be a helpful tool for regulating the DNA methylation of particular genes at the transcriptional level, which leads to the transcriptional silencing of various oncogenes (Jia et al. 2022). DNA methylation is highly interesting for therapeutic approaches since it is reversible, unlike genetic modifications. By binding to mRNAs, piRNAs may prevent the generation of cancer-related proteins at the post-transcriptional level. That is similar to the process by which miRNAs can function without Dicer. Interestingly, studies have shown that piRNAs can bind to other proteins in addition to PIWI proteins. Being able to bind proteins offers a way for therapeutic intervention. Since piRNAs are both a novel target for future therapeutic drug discovery or repurposed medicine targetingFootnote 3 and an efficient biomarker for cancer diagnosis, tumor gene therapy may advance due to ongoing research into specific interventions targeting piRNA expression. In both in vitro and in vivo studies, piRNA-targeted-therapy in conjunction with traditional chemotherapy has shown promising anticancer results. The combination of chemotherapeutic drugs such as paclitaxel, doxorubicin (DOX), and piR-54265 inhibitors in CRC or a piR-36712 analog in BC was more effective than treating with each agent alone (Mai et al. 2018; Tan et al. 2019).
Nano-bio-medicine (NBM) application
Recently, nanoparticle (NP) drug delivery systems (NDDSs) have become the most common system for co-delivering chemotherapy drugs and siRNA, which can inhibit the expression of genes associated with cancer or drug efflux transporters (Xiao et al. 2017). However, piRNA-based nano-formulations are still rarely used in the clinical field despite their promising potential application, and further studies are strongly required to develop new strategies to identify new barriers that hinder the use of RNA molecules in modern medicine. NPs can be classified into various classes according to their structure and features: polymeric, inorganic, lipid, and liposomal nanoparticles, and some of these types have proven to be convenient carriers for piRNAs (Han et al. 2024). The incorporation of specific piRNAs or piRNA inhibitors into the nanoparticle system has enabled their effective delivery to the targeted cells for the treatment of various cancer types. A nano-sized system of antago-miR-155 and piR-30074 with diethyl aminoethyl-dextran methyl methacrylate copolymer (DDMC) vector has achieved effective targeting and delivery of the bio-molecules to lung cancer cells, indicating a new therapeutic tool for lung cancer gene therapy (Klimenko 2017). Liposomal and polydopamine NPs of an encapsulated piR-1742 inhibitor suppressed the progression and metastasis of RCC. These NPs dramatically lowered the expression of MUC12 and piR-1742, implying the therapeutic potential of targeting piR-1742 in the treatment of RCC (Zhang et al. 2023). A magnetic NP-based gene therapy pre-loaded with anti-piR-823 in human BC cells transplanted in a mice xenograft model effectively suppressed carcinogenesis and tumor growth (Ding et al. 2021). The application of magnetic NP preloaded with piR-2158 in a BC xenograft model markedly inhibited the growth of mammary tumors, proliferation, and angiogenesis of breast cancer cells (Zhao et al. 2023). Gold nanoparticles (AuNPs) are inorganic nanoparticles most frequently used in various biomedical applications, including the detection and diagnostics of biomolecules. The AuNP system was developed as a fluorescent biosensing nano model to detect exosomal piR-823 ratiometrically in CRC samples (Sun et al. 2022). Taken together, treatment with a NP system pre-loaded with both chemotherapeutic drugs and piRNA mimic or inhibitor opens a new horizon in cancer therapy and is worth future exploration.
piR-823 contributing to cancer-hallmark
Cancer is often classified as a complex disease with multiple hallmarks, including cancer stem cells, resistance to apoptosis, angiogenesis, metastasis, tumor micro environment, chemoresistance, and cancer metabolism. As piRNAs are deregulated in virtually all human cancers, they show involvement in each of the cancer hallmarks as well. In this part, we describe the involvement of piRNAs especially piR-823 in cancer from a cancer hallmarks (Fig. 6).
piR-823 contribution in various cancer hallmarks. piR-823 involved in cancer biogenesis through regulating multiple cancer hallmarks such as cancer stem cell, cancer metabolism, tumor microenvironment, angiogenesis, metastasis, and apoptosis resistance (figure was created with BioRender.com). DNMT, DNA methyl transferase; TGF-1β, transforming growth factor-1beta; VEGF, vascular endothelial growth factor; CXCR4, C-X-C motif chemokine receptor 4; Bax, Bcl-2-associated X protein; Bcl2, B-cell lymphoma; WNT, wingless-related integration site; PINK1, PTEN-induced kinase1; MFN2, mitofusin2; G6PD, glucose-6-phosphate dehydrogenase; HIF‐1, hypoxia inducible factor-1; CSC, cancer stem cell
Cancer stem cells
With their ability to self-renew and differentiate, CSCs are a small and distinctive source of cells that can produce a wide range of tumor-forming cell types. They may also be the primary cause of drug resistance, metastasis, recurrence, and tumor progression (Zhao et al. 2023). Besides germline cells, piRNAs may have an extended function in regulating CSCs; however, the precise functions of piRNAs remain mostly unknown (Huang et al. 2013b). Through the upregulation of piR-823 expression and the activation of DNMT3B, G-MDSCs have provided MM cells with stem-like characteristics. The elevated levels of CSC-related genes and proteins in MM cells caused by G-MDSCs are reversed by piR-823 silencing. Considering this, piR-823 appears to be an essential mediator in the maintenance of MM stem cells (Ai et al. 2019). Furthermore, piR-823 controlled DNA methylation to promote stemness and proliferation of breast CSCs, indicating that DNA methylation patterns are a mechanism for piRNA-mediated carcinogenesis and offer an in-depth comprehension of the various epigenetic patterns of stem cells and somatic cells (Ding et al. 2021).
Resistance to apoptosis
Resistance to apoptosis is one of the major cancer hallmarks contributing to tumor occurrence, progression, and chemoresistance (Modabber et al. 2023; Mohammad et al. 2015). In recent years, PIWI/piRNAs have taken part in the process of apoptosis (Tan et al. 2022). piR-823 regulates apoptosis in a diversity of cells. In MM, the overexpression of piR-823 significantly attenuates apoptosis through inhibition of caspase-3 activation, downregulation of Bcl-2-associated X protein (Bax), and upregulation of Bcl2 (Yan et al. 2015). The two most significant mitophagy receptors, parkin and PINK1, are tumor suppressor genes involved in inducing apoptosis, establishing a significant association between mitophagy pathways and apoptosis (Bernardini et al. 2017). Accordingly, inhibiting piR-823 promoted apoptosis in CRC cell lines in a way dependent on mitophagy. In piR-823-silenced cells, cleaved PARP, cleaved caspase-9, and cleaved caspase-3 protein levels decreased due to parkin inhibition (Wang et al. 2022b). Inhibiting piR-823 in CRC tissues induces cell apoptosis, causes cell cycle arrest in the G1 phase, and reduces the progression of CRC (Yin et al. 2017). Various studies have shown that the TGF-1β signaling pathway plays a key role in the cell apoptosis. The combination of piR-823 and EIF3B-mediated TGF-1β expression in HSC promotes liver fibrosis and cirrhosis (Tang et al. 2018).
Tumor metastasis and invasion
The primary feature of malignant tumors is metastasis, which is also a crucial indicator of tumor progression and a poor outcome. DNA methylation, histone alteration, and piRNA aberrant expression are among the epigenetic changes attributed to cancer metastasis. Comprehending the piRNA-related epigenetic pathways of tumor metastasis might help in the finding of novel tumor markers and therapeutic interventions (Liu et al. 2018a). By comparing benign, metastatic, and non-metastatic kidney tissue, 19 piRNAs were identified to be differentially expressed, and 46 were substantially related to metastasis. Three piRNAs (piR-32051, piR-39894, and piR-43607), among the metastasis-related piRNAs, were associated with metastatic renal carcinoma of advanced tumor stage and cancer-specific characteristics in patients (Li et al. 2015). piR-823 and PIWI protein expression levels have been correlated to TNM staging in renal cancer (Slaby et al. 2016). Furthermore, the TNM stage and distant metastases in GC were positively associated with the piR-823 level (Cui et al. 2011). Consequently, the prognosis of the disease and the TNM stage can be predicted using piRNAs as biomarkers. Moreover, piRNAs may act as a switch that permits tumor proliferation, spread, and metastasis.
Tumor angiogenesis
Angiogenesis is a critical phase in the development and progression of tumors, which promotes invasive tumor proliferation and distant metastasis (Teleanu et al. 2019). Only a few studies explored the role of piRNAs, specifically piR-823, in tumor angiogenesis (Chen et al. 2021). In MM cells, the VEGF pathway may have a paracrine angiogenic role that promotes the development of tumors. piR-823 could promote carcinogenesis in MM by controlling the release of VEGF (Yan et al. 2015).
Tumor microenvironment (TME)
Because the tumor microenvironment offers a rich environment for exploring novel therapeutic targets, it is essential to the invasion and metastasis of tumors (Elanany et al. 2023). Tumor cells may create such as microvesicles and exosomes to interact with other stromal cells by transferring ncRNAs that has the potential to alter the tumor microenvironment and promote tumor growth (Zhu et al. 2019). A favorable environment for MM may be created by piR-823 supplied by MM-derived EVs, which may improve the survival and angiogenic function of the ECs (Li et al. 2019).
Cancer metabolism
Another apparent cancer hallmark is the way energy is metabolized in cancer cells. According to recent research, cancer cells can modify their glucose metabolism and shift toward aerobic glycolysis by increasing glycolysis in the presence of oxygen and inhibiting the tricarboxylic acid cycle (Pavlova and Thompson 2016). Thus, key features of human cancer that distinguish it apart from normal tissues include excessive energy production and metabolic reprogramming, which are controlled by several oncogenes and tumor suppressors (Dong et al. 2020). The suppression of HIF-1α ubiquitination by upregulating G6PD expression can be achieved by upregulating piR-823 in CRC cells. This can lead to an increase in glucose uptake and a decrease in intracellular ROS levels in cancer cells (Yin et al. 2017). A novel type of pro-inflammatory-programmed cell death known as hyperglycemic-induced pyroptosis can be inhibited by upregulating piRNA-823 expression in endothelial cells (Tan et al. 2024).
Future prospective from the expert opinion perspective
Besides all the addressed research gaps, the current comprehensive information about piRNA, particularly piR-823, in various tumors opens the door for future prospective studies experimentally via knocking down piR-823 and its axis of related genes to prove the concept of utilizing piRNA directly to inhibit the transcription of a specific down-stream gene or multiple genes or proteins, mediated through other epigenetic alteration(s). More thorough research at the cellular and animal levels on the precise mechanism by which piRNA identifies its target in the genome and the possibility of off-target effects would be required to accomplish such an objective (Sun and Han 2019). Further studies are required to explore the relation of piR-823 with adipokines SNPs (Aboouf et al. 2015), or adipokines expression (El-Mesallamy et al. 2013), inflammation molecular markers, namely, the tumor immune microenvironment, insulin resistance or insulin-like growth factors (El-Mesallamy et al. 2012), and related hormones or genes in various cancer types. Moreover, future research to link piRNA SNP variants or haplotypes in different types of cancer will help identify clinical cancer cases at an early stage or low grade, ensuring personalized earlier diagnosis for better reflection on improving patients’ survival. Consequently, a step toward ncRNA precision is to implement ncRNA measurements in liquid biopsy or tumor tissues in addition to the epigenome project.
Limitations
Compared to miRNAs, using piRNAs in the clinical field is still in its infancy, with limited information about piRNAs’ role in various non-communicable diseases, including different cancer types or rare diseases. Moreover, the possibility that many piRNAs could originate from the same genomic region, known as the piRNA cluster, adds another layer of difficulty to the mechanistic investigation of piRNAs; therefore, examining such clusters is a necessity.
Conclusion(s)
It should not be too long until piRNAs are introduced into diagnostic, prognostic, and therapeutic clinical applications, given the amount of data that has been obtained regarding piRNAs for the sake of ncRNA use in precision medicine. piR-823 has become a promising cancer-specific biomarker due to its aberrant expression in several cancers. However, further research is still needed to fully understand the tumor-suppressive or carcinogenic functions of piR-823. Identifying the molecular mechanistic functions of piR-823 in various cancer hallmarks is likely to provide more insights into their roles in cancer initiation, progression, and metastasis. Interestingly, the presence of piRNAs in the exosomes suggests the potential application of piRNAs for distant target sites delivered along with drugs via exosomes or nano-biomedicine.
Data availability
No datasets were generated or analysed during the current study.
Notes
research gap no. 1.
research gap no. 2.
research gap no. 3.
Abbreviations
- 3′ UTR:
-
3′ Untranslated region
- Ago:
-
Argonaute3
- Aub:
-
Aubergine
- BC:
-
Breast cancer
- Bcl:
-
B-cell lymphoma
- CDK:
-
Cyclin-dependent kinase
- CDS:
-
Protein-coding sequence
- CEA:
-
Carcinoembryonic antigen
- CSCs:
-
Cancer stem cells
- DNMT:
-
DNA methyl transferase
- DOX:
-
Doxorubicin
- ECs:
-
Endothelial cells
- EIF3B:
-
Eukaryotic initiation factor 3 B
- ER:
-
Estrogen receptor
- EVs:
-
Extracellular vehicles
- G-MDSCs:
-
Granulocytic-MDSCs
- G6PD:
-
Glucose-6-phosphate dehydrogenase
- GC:
-
Gastric cancer
- GI:
-
Gastrointestinal
- GWAS:
-
Genome-wide association study
- HCC:
-
Hepatocellular carcinoma
- Her2:
-
Human epidermal growth factor receptor 2
- HIF-1 α:
-
Hypoxia-inducible factor-1-alpha
- HSCs:
-
Hepatic stellate cells
- HSF1:
-
Heat shock factor 1
- LncRNA:
-
Long non-coding RNA
- MDSCs:
-
Myeloid-derived suppressor cells
- miRNA:
-
MicroRNA
- MM:
-
Multiple myeloma
- MMP:
-
Matrix metallopeptidase
- mRNA:
-
Messenger RNA
- mTOR:
-
Mammalian target of rapamycin
- ncRNA:
-
Non-coding RNA
- NP:
-
Nanoparticle
- nt:
-
Nucleotide
- PCR:
-
Polymerase chain reaction
- PINK:
-
PTEN-induced kinase1
- piRNAs:
-
PIWI-interacting RNAs
- PIWIL:
-
PIWI-like protein
- PR:
-
Progesterone receptor
- RCC:
-
Renal cell carcinoma
- RISC:
-
RNA-induced silencing complex
- ROS:
-
Reactive oxygen species
- siRNA:
-
Small interfering RNA
- SNP:
-
Single-nucleotide polymorphism
- STAT3:
-
Signal transducer and activator of transcription 3
- TEs:
-
Transposable elements
- TGF-β:
-
Transforming growth factor beta
- TGS:
-
Transcriptional gene silencing
- TNM:
-
Tumor node metastasis
- VEGF:
-
Vascular endothelial growth factor
References
Abaza T, Abd MK, El-Aziz KA, Daniel PK, Papatsirou M, Fahmy SA, Hamdy NM, Kontos CK, Youness RA (2023) Emerging role of circular RNAs in hepatocellular carcinoma immunotherapy. Int J Mol Sci 24(22):16484. https://doi.org/10.3390/ijms242216484
Abd El Fattah YK, Abulsoud AI, Abdelhamid SG, Abdelhalim S, Hamdy NM (2023) CCDC144NL-AS1/hsa-miR-143–3p/HMGA2 interaction: In-silico and clinically implicated in CRC progression, correlated to tumor stage and size in case-controlled study; step toward ncRNA precision. Int J Biol Macromol 253(Pt 2):126739. https://doi.org/10.1016/j.ijbiomac.2023.126739
Aboouf MA, Hamdy NM, Amin AI, El-Mesallamy HO (2015) Genotype screening of APLN Rs3115757 variant in Egyptian women population reveals an association with obesity and insulin resistance. Diabetes Res Clin Pract 109(1):40–47. https://doi.org/10.1016/j.diabres.2015.05.016
Ai L, Mu S, Sun C, Fan F, Yan H, Qin Y, Cui G, Wang Y, Guo T, Mei H, Wang H, Yu H (2019) Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing PiRNA-823 expression and DNMT3B activation. Mol Cancer 18(1):88. https://doi.org/10.1186/s12943-019-1011-5
Ameli Mojarad M, Mojarad MA, Shojaee B, Nazemalhosseini-Mojarad E (2022) PiRNA: a promising biomarker in early detection of gastrointestinal cancer. Pathol - Res Pract 230:153757. https://doi.org/10.1016/j.prp.2021.153757
Anon (n.d.-a) PiRNAdb https://Www.Pirnadb.Org/
Anon (n.d.-b) PiRPheno: http://Www.Biomedical-Web.Com/Pirpheno/
Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Jingyue Ju, Sheridan R, Sander C, Zavolan M, Tuschl T (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442(7099):203–207. https://doi.org/10.1038/nature04916
Assumpção CB, Calcagno DQ, Araújo TMT, Batista SE, dos Santos Â, Ribeiro KC, dos Santos G, Riggins J, Burbano RR, Assumpção PP (2015) The role of PiRNA and its potential clinical implications in cancer. Epigenomics 7(6):975–984. https://doi.org/10.2217/epi.15.37
Barckmann B, Pierson S, Dufourt J, Papin C, Armenise C, Port F, Grentzinger T, Chambeyron S, Baronian G, Desvignes J-P, Curk T, Simonelig M (2015) Aubergine ICLIP reveals PiRNA-dependent decay of MRNAs involved in germ cell development in the early embryo. Cell Rep 12(7):1205–1216. https://doi.org/10.1016/j.celrep.2015.07.030
Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D (2016) Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell 30(6):836–848. https://doi.org/10.1016/j.ccell.2016.10.009
Bernardini JP, Lazarou M, Dewson G (2017) Parkin and mitophagy in cancer. Oncogene 36(10):1315–1327. https://doi.org/10.1038/onc.2016.302
Chalbatani GM, Dana H, Memari F, Gharagozlou E, Ashjaei S, Kheirandish P, Marmari V, Mahmoudzadeh H, Mozayani F, Maleki AR, Sadeghian E, Nia EZ, Miri SR, zainali Nia N, Rezaeian O, Eskandary A, Razavi N, Shirkhoda M, Rouzbahani FN (2019) Biological function and molecular mechanism of PiRNA in cancer. Pract Lab Med 13:e00113. https://doi.org/10.1016/j.plabm.2018.e00113
Chen S, Ben S, Xin J, Li S, Zheng R, Wang H, Fan L, Du M, Zhang Z, Wang M (2021) The biogenesis and biological function of PIWI-interacting RNA in cancer. J Hematol Oncol 14(1):93. https://doi.org/10.1186/s13045-021-01104-3
Cheng J, Guo J-M, Xiao B-X, Miao Y, Jiang Z, Zhou H, Li Q-N (2011) PiRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta 412(17–18):1621–1625. https://doi.org/10.1016/j.cca.2011.05.015
Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J (2012) PiR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett 315(1):12–17. https://doi.org/10.1016/j.canlet.2011.10.004
Cheng Y, Wang Q, Jiang W, Bian Y, Zhou Y, Gou A, Zhang W, Fu K, Shi W (2019) Emerging roles of PiRNAs in cancer: challenges and prospects. Aging 11(21):9932–46. https://doi.org/10.18632/aging.102417
Chu H, Xia L, Qiu X, Du D, Zhu L, Jin J, Hui G, Hua Q, Du M, Tong N, Chen J, Zhang Z, Wang M (2015) Genetic variants in noncoding PIWI-interacting RNA and colorectal cancer risk. Cancer 121(12):2044–2052. https://doi.org/10.1002/cncr.29314
Cui L, Lou Y, Zhang X, Zhou H, Deng H, Song H, Y X, Xiao B, Wang W, Guo J (2011) Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using PiRNAs as markers. Clin Biochem 44(13):1050–1057. https://doi.org/10.1016/j.clinbiochem.2011.06.004
Czech B, Hannon GJ (2016) One loop to rule them all: the ping-pong cycle and PiRNA-guided silencing. Trends Biochem Sci 41(4):324–337. https://doi.org/10.1016/j.tibs.2015.12.008
Ding X, Li Y, Lü J, Zhao Q, Guo Y, Lu Z, Ma W, Liu P, Pestell RG, Liang C, Yu Z (2021) PiRNA-823 is involved in cancer stem cell regulation through altering DNA methylation in association with luminal breast cancer. Front Cell Dev Biol 9. https://doi.org/10.3389/fcell.2021.641052
Dong Y, Tu R, Liu H, Qing G (2020) Regulation of cancer cell metabolism: oncogenic MYC in the driver’s seat. Signal Transduct Target Ther 5(1):124. https://doi.org/10.1038/s41392-020-00235-2
Elanany MM, Mostafa D, Hamdy NM (2023) Remodeled tumor immune microenvironment (TIME) parade via natural killer cells reprogramming in breast cancer. Life Sci 2023(330):121997
El-Aziz MK, Abd AD, Kiriacos CJ, Fahmy SA, Hamdy NM, Youness RA (2023) Decoding hepatocarcinogenesis from a noncoding RNAs perspective. J Cell Physiol 238(9):1982–2009. https://doi.org/10.1002/jcp.31076
El-Derany MO, Hamdy NM, Al-Ansari NL, El-Mesallamy HO (2016) Integrative role of vitamin D related and interleukin-28B genes polymorphism in predicting treatment outcomes of chronic hepatitis C. BMC Gastroenterol 16(1):19. https://doi.org/10.1186/s12876-016-0440-5
Eldosoky M, Hammad R, Elmadbouly A, Aglan R, AbdelHamid S, Alboraie M, Hassan D, Shaheen M, Rushdi A, Ahmed R, Abdelbadea A, Abdelmageed N, Elshafei A, Ali E, Abo-Elkheir O, Zaky S, Hamdy N, Lambert C (2023) Diagnostic significance of Hsa-MiR-21-5p, Hsa-MiR-192-5p, Hsa-MiR-155-5p, Hsa-MiR-199a-5p panel and ratios in hepatocellular carcinoma on top of liver cirrhosis in HCV-infected patients. Int J Mol Sci 24(4):3157. https://doi.org/10.3390/ijms24043157
El-Mesallamy HO, Hamdy NM, Zaghloul AS, Sallam AM (2012) Serum retinol binding protein-4 and neutrophil gelatinase-associated lipocalin are interrelated in pancreatic cancer patients. Scand J Clin Lab Invest 72(8):602–607. https://doi.org/10.3109/00365513.2012.723135
El-Mesallamy HO, Hamdy NM, Zaghloul AS, Sallam AM (2013) Clinical value of circulating lipocalins and insulin-like growth factor axis in pancreatic cancer diagnosis. Pancreas 42(1):149–154. https://doi.org/10.1097/MPA.0b013e3182550d9d
El-Sheikh NM, Abulsoud AI, Fawzy A, Wasfey EF, Hamdy NM (2023) LncRNA NNT-AS1/Hsa-MiR-485–5p/HSP90 axis in-silico and clinical prospect correlated-to histologic grades-based CRC stratification: a step toward NcRNA precision. Pathol - Rese Pract 247:154570. https://doi.org/10.1016/j.prp.2023.154570
Emam O, Wasfey EF, Hamdy NM (2022) Notch-associated LncRNAs profiling circuiting epigenetic modification in colorectal cancer. Cancer Cell Int 22(1):316. https://doi.org/10.1186/s12935-022-02736-2
Feldman BJ, Feldman D (2001) The development of androgen-independent prostate cancer. Nat Rev Cancer 1(1):34–45. https://doi.org/10.1038/35094009
Feng J, Yang M, Wei Q, Song F, Zhang Y, Wang X, Liu B, Li J (2020) Novel evidence for oncogenic PiRNA-823 as a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. J Cell Mol Med 24(16):9028–9040. https://doi.org/10.1111/jcmm.15537
Fu A, Jacobs DI, Hoffman AE, Zheng T, Zhu Y (2015) PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis 36(10):1094–1102. https://doi.org/10.1093/carcin/bgv105
Furukawa Y, Kikuchi J (2015) Molecular pathogenesis of multiple myeloma. Int J Clin Oncol 20(3):413–422. https://doi.org/10.1007/s10147-015-0837-0
Goh T-X, Tan S-L, Roebuck MM, Teo S-H, Kamarul T (2022) A systematic review of extracellular vesicle-derived Piwi-interacting RNA in human body fluid and its role in disease progression. Tissue Eng C: Methods 28(10):511–528. https://doi.org/10.1089/ten.tec.2022.0092
Gu X, Wang C, Deng H, Qing C, Liu R, Liu S, Xue X (2020) Exosomal PiRNA profiling revealed unique circulating PiRNA signatures of cholangiocarcinoma and gallbladder carcinoma. Acta Biochim Biophys Sin 52(5):475–484. https://doi.org/10.1093/abbs/gmaa028
Guo B, Li D, Du L, Zhu X (2020) PiRNAs: Biogenesis and their potential roles in cancer. Cancer Metastasis Rev 39(2):567–575. https://doi.org/10.1007/s10555-020-09863-0
Hammad R, Eldosoky MA, Elmadbouly AA, Aglan RB, AbdelHamid SG, Zaky S, Ali E, Hakam F-Z, Mosaad AM, Abdelmageed NA, Kotb FM, Kotb HG, Hady AA, Abo-Elkheir OI, Kujumdshiev S, Sack U, Lambert C, Hamdy NM (2023) Monocytes subsets altered distribution and dysregulated plasma Hsa-MiR-21-5p and Hsa-MiR-155-5p in HCV-linked liver cirrhosis progression to hepatocellular carcinoma. J Cancer Res Clin Oncol 149(17):15349–15364. https://doi.org/10.1007/s00432-023-05313-w
Han BW, Zamore PD (2014) PiRNAs. Curr Biol 24(16):R730–R733. https://doi.org/10.1016/j.cub.2014.07.037
Han H, Fan G, Song S, Jiang Y, Qian C, Zhang W, Su Q, Xue X, Zhuang W, Li B (2021) PiRNA-30473 contributes to tumorigenesis and poor prognosis by regulating M6A RNA methylation in DLBCL. Blood 137(12):1603–14. https://doi.org/10.1182/blood.2019003764
Han R, Rao X, Zhou H, Lu L (2024) Synergistic immunoregulation: harnessing CircRNAs and PiRNAs to amplify PD-1/PD-L1 inhibition therapy. Int J Nanomed 19:4803–4834. https://doi.org/10.2147/IJN.S461289
Hanusek K, Poletajew S, Kryst P, Piekiełko-Witkowska A, Bogusławska J (2022) PiRNAs and PIWI proteins as diagnostic and prognostic markers of genitourinary cancers. Biomolecules 12(2):186. https://doi.org/10.3390/biom12020186
Henaoui IS, Jacovetti C, Mollet IG, Guay C, Sobel J, Eliasson L, Regazzi R (2017) PIWI-interacting RNAs as novel regulators of pancreatic beta cell function. Diabetologia 60(10):1977–1986. https://doi.org/10.1007/s00125-017-4368-2
Huang G, Hu H, Xue X, Shen S, Gao E, Guo G, Shen X, Zhang X (2013a) Altered expression of PiRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol 15(7):563–568. https://doi.org/10.1007/s12094-012-0966-0
Huang T, Alvarez A, Bo Hu, Cheng S-Y (2013b) Noncoding RNAs in cancer and cancer stem cells. Chin J Cancer 32(11):582–593. https://doi.org/10.5732/cjc.013.10170
Huang H, Li Y, Szulwach KE, Zhang G, Jin P, Chen D (2014a) AGO3 slicer activity regulates mitochondria–nuage localization of armitage and PiRNA amplification. J Cell Biol 206(2):217–230. https://doi.org/10.1083/jcb.201401002
Huang Y, Bai JY, Ren HT (2014b) PiRNA biogenesis and its functions. Russ J Bioorg Chem 40(3):293–299. https://doi.org/10.1134/S1068162014030169
Iliev R, Fedorko M, Machackova T, Mlcochova H, Svoboda M, Pacik D, Dolezel J, Stanik M, Slaby O (2016) Expression levels of PIWI-interacting RNA, PiR-823, are deregulated in tumor tissue, blood serum and urine of patients with renal cell carcinoma. Anticancer Res 36(12):6419–24. https://doi.org/10.21873/anticanres.11239
Jacobs DI, Qin Q, Lerro MC, Alan Fu, Dubrow R, Claus EB, DeWan AT, Wang G, Lin H, Zhu Y (2016) PIWI-interacting RNAs in gliomagenesis: evidence from post-GWAS and functional analyses. Cancer Epidemiol Biomark Prev 25(7):1073–1080. https://doi.org/10.1158/1055-9965.EPI-16-0047
Jeong H, Park KH, Lee Y, Jeong A, Choi S, Kim KW (2021) The regulation and role of PiRNAs and PIWI proteins in cancer. Processes 9(7):1208. https://doi.org/10.3390/pr9071208
Jia D-D, Jiang H, Yi-Fei Zhang Yu, Zhang L-L, Zhang Y-F (2022) The regulatory function of PiRNA/PIWI complex in cancer and other human diseases: the role of DNA methylation. Int J Biol Sci 18(8):3358–3373. https://doi.org/10.7150/ijbs.68221
Jiang B-R, Wu W-Y, Chien C-H, Tsai JJP, Chan W-L (2016) PiRNAtarget: The integrated database for mining functionality of PiRNA and its targets. In: 2016 IEEE 16th Int Conf Bioinforma Bioeng (BIBE). IEEE, pp 382–386
Kelley D, Rinn J (2012) Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol 13(11):R107. https://doi.org/10.1186/gb-2012-13-11-r107
Khalili P, Maddah R, Maleknia M, Amiri BS, Forouzani F, Hasanvand A, Rezaeeyan H (2023) Evaluation of genes and molecular pathways involved in the progression of monoclonal gammopathy of undetermined significance (MGUS) to multiple myeloma: a systems biology approach. Mol Biotechnol 65(8):1275–1286. https://doi.org/10.1007/s12033-022-00634-6
Klimenko OV (2017) Joint action of the nano-sized system of small non-coding RNAs with DDMC vector and recombinant IL-7 reprograms A-549 lung adenocarcinoma cells into CD4+ cells. Immunother Open Access 03(01). https://doi.org/10.4172/2471-9552.1000137
Korn C, Méndez-Ferrer S (2017) Myeloid malignancies and the microenvironment. Blood 129(7):811–822. https://doi.org/10.1182/blood-2016-09-670224
Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li En, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T (2008) DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22(7):908–917. https://doi.org/10.1101/gad.1640708
Li P-F (2014) Non-coding RNAs and gastric cancer. World J Gastroenterol 20(18):5411. https://doi.org/10.3748/wjg.v20.i18.5411
Li Y, Wu X, Gao H, Jin JM, Li AX, Kim YS, Pal SK, Nelson RA, Lau CM, Guo C, Mu B, Wang J, Wang F, Wang J, Zhao Y, Chen W, Rossi JJ, Weiss LM, Wu H (2015) Piwi-interacting RNAs (PiRNAs) are dysregulated in renal cell carcinoma and associated with tumor metastasis and cancer-specific survival. Mol Med 21(1):381–388. https://doi.org/10.2119/molmed.2014.00203
Li B, Hong J, Hong M, Wang Y, Yu T, Zang S, Wu Q (2019) PiRNA-823 delivered by multiple myeloma-derived extracellular vesicles promoted tumorigenesis through re-educating endothelial cells in the tumor environment. Oncogene 38(26):5227–5238. https://doi.org/10.1038/s41388-019-0788-4
Li Y, Wang Z, Ajani JA, Song S (2021) Drug resistance and cancer stem cells. Cell Commun Signal 19(1):19. https://doi.org/10.1186/s12964-020-00627-5
Li G, Yi X, Du S, Gong L, Wu Q, Cai J, Sun S, Cao Y, Chen L, Xu L, Wang Z (2023) Tumour-derived exosomal PiR-25783 promotes omental metastasis of ovarian carcinoma by inducing the fibroblast to myofibroblast transition. Oncogene 42(6):421–433. https://doi.org/10.1038/s41388-022-02560-y
Li Y, Dong Y, Zhao S, Gao J, Hao X, Wang Z, Li M, Wang M, Liu Y, Yu X, Xu W (2022) Serum-derived PiR-Hsa-164586 of extracellular vesicles as a novel biomarker for early diagnosis of non-small cell lung cancer. Front Oncol 12. https://doi.org/10.3389/fonc.2022.850363
Lin X, Xia Y, Dan Hu, Mao Q, Zongyang Yu, Zhang H, Li C, Chen G, Liu F, Zhu W, Shi Yi, Zhang H, Zheng J, Sun T, Jianying Xu, Chao H, Zheng X, Luο X (2019) Transcriptome-wide PiRNA profiling in human gastric cancer. Oncol Rep. https://doi.org/10.3892/or.2019.7073
Liu J, Zhang S, Cheng B (2018a) Epigenetic roles of PIWI-interacting RNAs (PiRNAs) in cancer metastasis (Review). Oncol Rep. https://doi.org/10.3892/or.2018.6684
Liu Y, Zhang J, Li A, Liu Z, He Z, Yuan X, Tuo S (2018b) Prediction of cancer-associated PiRNA–MRNA and PiRNA–LncRNA interactions by integrated analysis of expression and sequence data. Tsinghua Sci Technol 23(2):115–25. https://doi.org/10.26599/TST.2018.9010056
Liu Y, Dou M, Song X, Dong Y, Liu S, Liu H, Tao J, Li W, Yin X, Xu W (2019) The emerging role of the PiRNA/Piwi complex in cancer. Mol Cancer 18(1):123. https://doi.org/10.1186/s12943-019-1052-9
Liu Y, Li A, Xie G, Liu G, Hei X (2021) Computational methods and online resources for identification of PiRNA-related molecules. Interdiscip Sci: Comput Life Sci 13(2):176–191. https://doi.org/10.1007/s12539-021-00428-5
Liu Y, Li A, Zhu Y, Pang X, Hei X, Xie G, Wu F-X (2023) PiRSNP: a database of PiRNA-related SNPs and their effects on cancer related PiRNA functions. Curr Bioinform 18(6):509–516. https://doi.org/10.2174/1574893618666230320144630
Ma H, Wang H, Tian F, Zhong Y, Liu Z, Liao A (2020) PIWI-interacting RNA-004800 is regulated by S1P receptor signaling pathway to keep myeloma cell survival. Front Oncol 10. https://doi.org/10.3389/fonc.2020.00438
Mahmoud MM, Sanad EF, Elshimy RAA, Hamdy NM (2021a) Competitive endogenous role of the LINC00511/MiR-185–3p axis and MiR-301a-3p from liquid biopsy as molecular markers for breast cancer diagnosis. Front Oncol 11. https://doi.org/10.3389/fonc.2021.749753
Mahmoud MM, Sanad EF, Hamdy NM (2021b) MicroRNAs’ role in the environment-related non-communicable diseases and link to multidrug resistance, regulation, or alteration. Environ Sci Pollut Res 28(28):36984. https://doi.org/10.1007/s11356-021-14550-w
Mai D, Ding P, Tan L, Zhang J, Pan Z, Bai R, Li C, Li M, Zhou Y, Tan W, Zhou Z, Li Y, Zhou A, Ye Y, Pan L, Zheng Y, Su J, Zuo Z, Liu Z, Zhao Q, Li X, Huang X, Li W, Wu S, Jia W, Zou S, Wu C, R-h Xu, Zheng J, Lin D (2018) PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics 8(19):5213–5230. https://doi.org/10.7150/thno.28001
Mei Y, Clark D, Mao L (2013) Novel dimensions of PiRNAs in cancer. Cancer Lett 336(1):46–52. https://doi.org/10.1016/j.canlet.2013.04.008
Modabber N, Mahboub SS, Khoshravesh S, Karimpour F, Karimi A, Goodarzi V (2023) Evaluation of long non-coding RNA (LncRNA) in the pathogenesis of chemotherapy resistance in cervical cancer: diagnostic and prognostic approach. Mol Biotechnol. https://doi.org/10.1007/s12033-023-00909-6
Mohammad RM, Muqbil I, Lowe L, Yedjou C, Hsu H-Y, Lin L-T, Siegelin MD, Fimognari C, Kumar NB, Ping Dou Q, Yang H, Samadi AK, Russo GL, Spagnuolo C, Ray SK, Chakrabarti M, Morre JD, Coley HM, Honoki K, Fujii H, Georgakilas AG, Amedei A, Niccolai E, Amin A, Ashraf SS, Helferich WG, Yang X, Boosani CS, Guha G, Bhakta D, Ciriolo MR, Aquilano K, Chen S, Mohammed SI, Nicol Keith W, Bilsland A, Halicka D, Nowsheen S, Azmi AS (2015) Broad targeting of resistance to apoptosis in cancer. Semin Cancer Biol 35:S78-103. https://doi.org/10.1016/j.semcancer.2015.03.001
Mojarad MA, Mojarad MA (2021) PiRNAs and PIWI proteins as potential biomarkers in colorectal cancer. J Clin Images Med Case Rep 2(6). https://doi.org/10.52768/2766-7820/1447
Moyano M, Stefani G (2015) PiRNA involvement in genome stability and human cancer. J Hematol Oncol 8(1):38. https://doi.org/10.1186/s13045-015-0133-5
Muhammad A, Waheed R, Khan NA, Jiang H, Song X (2019) PiRDisease v1.0: a manually curated database for PiRNA associated diseases. Database 2019. https://doi.org/10.1093/database/baz052
Öner Ç, Coşan DT, Çolak E (2016) Estrogen and androgen hormone levels modulate the expression of PIWI interacting RNA in prostate and breast cancer. PLoS ONE 11(7):e0159044. https://doi.org/10.1371/journal.pone.0159044
Öner Ç, Soyergin D, Özyurt A, Çolak E (2022) 4-Hydroxycoumarin effects on both cellular and genetic characteristics of hepatocellular carcinoma cells. Cytol Genet 56(3):292–300. https://doi.org/10.3103/S0095452722030094
Oner C (2021) Genetic markers indicate that 1,25-dihydroxyvitamin D treatment may not protect against hepatocellular carcinoma. Eurasian J Med Oncol. https://doi.org/10.14744/ejmo.2021.65874
Oner C (2022) PIWI interacting RNA-823: epigenetic regulator of the triple negative breast cancer cells proliferation. Eurasian J Med Oncol. https://doi.org/10.14744/ejmo.2022.12505
Ou B, Liu Y, Gao Z, Xu J, Yan Y, Li Y, Zhang J (2022) Senescent neutrophils-derived exosomal PiRNA-17560 promotes chemoresistance and EMT of breast cancer via FTO-mediated M6A demethylation. Cell Death Dis 13(10):905. https://doi.org/10.1038/s41419-022-05317-3
Pavlova NN, Thompson CB (2016) The emerging hallmarks of cancer metabolism. Cell Metab 23(1):27–47. https://doi.org/10.1016/j.cmet.2015.12.006
Peng L, Song L, Liu C, Lv X, Li X, Jie J, Zhao D, Li D (2016) PiR-55490 inhibits the growth of lung carcinoma by suppressing MTOR signaling. Tumor Biology 37(2):2749–2756. https://doi.org/10.1007/s13277-015-4056-0
Raposo G, Stoorvogel W (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200(4):373–383. https://doi.org/10.1083/jcb.201211138
Rezaeean H, Kaydani GA, Saki N, Razmjoo S, Labibzadeh M, Yaghooti H (2021) The IFN-Ɣ + 874 A/T polymorphism is associated with malignant breast cancer in a population from the Southwest of Iran. BMC Res Notes 14(1):147. https://doi.org/10.1186/s13104-021-05543-6
Rizk NI, Kassem DH, Abulsoud AI, ..., Kamal MM, Hamdy NM (2024) “Revealing the role of serum exosomal novel long non-coding RNA NAMPT-AS as a promising diagnostic/prognostic biomarker in colorectal cancer patients.” Life Sci 352:122850
Rizzo F, Rinaldi A, Marchese G, Coviello E, Sellitto A, Cordella A, Giurato G, Nassa G, Ravo M, Tarallo R, Milanesi L, Destro A, Torzilli G, Roncalli M, Di Tommaso L, Weisz A (2016) Specific patterns of PIWI-interacting small noncoding RNA expression in dysplastic liver nodules and hepatocellular carcinoma. Oncotarget 7(34):54650–61. https://doi.org/10.18632/oncotarget.10567
Robak P, Drozdz I, Szemraj J, Robak T (2018) Drug resistance in multiple myeloma. Cancer Treat Rev 70:199–208. https://doi.org/10.1016/j.ctrv.2018.09.001
Roy J, Anand K, Mohapatra S, Nayak R, Chattopadhyay T, Mallick B (2020) Single nucleotide polymorphisms in PiRNA-pathway genes: an insight into genetic determinants of human diseases. Mol Genet Genomics 295(1):1–12. https://doi.org/10.1007/s00438-019-01612-5
Rui T, Wang K, Xiang A, Guo J, Tang N, Jin X, Lin Y, Liu J, Zhang X (2023) Serum exosome-derived PiRNAs could be promising biomarkers for HCC diagnosis. Int J Nanomed 18:1989–2001. https://doi.org/10.2147/IJN.S398462
Sabbah NA, Abdalla WM, Mawla WA, AbdAlMonem N, Gharib AF, Abdul-Saboor A, Abdelazem AS, Raafat N (2021) PiRNA-823 is a unique potential diagnostic non-invasive biomarker in colorectal cancer patients. Genes 12(4):598. https://doi.org/10.3390/genes12040598
Salama S, Hamdy N, El-shemy R, El-Mesallamy H (2021) Clinical significance of the transcription factor SOX11, cell-cell adhesion protein E-cadherin and zinc finger protein BCL11A in the diagnosis of breast cancer. Arch Pharm Sci Ain Shams Univ 5(1):97–110. https://doi.org/10.21608/aps.2021.75847.1058
Sarkar A, Maji RK, Saha S, Ghosh Z (2014) PiRNAQuest: searching the PiRNAome for silencers. BMC Genomics 15(1):555. https://doi.org/10.1186/1471-2164-15-555
Slaby O, Iliev R, Stanik M, Fedorko M, Poprach A, Vychytilova-Faltejskova P, Slaba K, Svoboda M, Fabian P, Pacik D, Dolezel J (2016) Decreased expression levels of PIWIL1, PIWIL2, and PIWIL4 are associated with worse survival in renal cell carcinoma patients. OncoTargets Ther 217. https://doi.org/10.2147/OTT.S91295
Su J-F, Zhao F, Gao Z-W, Hou Y-J, Li Y-Y, Duan L-J, Lun S-M, Yang H-J, Li J-K, Dai N-T, Shen F-F, Zhou F-Y (2020) PiR-823 demonstrates tumor oncogenic activity in esophageal squamous cell carcinoma through DNA methylation induction via DNA methyltransferase 3B. Pathol - Res Pract 216(4):152848. https://doi.org/10.1016/j.prp.2020.152848
Sun T, Han X (2019) The disease-related biological functions of PIWI-interacting RNAs (PiRNAs) and underlying molecular mechanisms. ExRNA 1(1):21. https://doi.org/10.1186/s41544-019-0021-1
Sun Z, Tong Y, Li J, Wang Y, Gao F, Li H, Wang C, Du L, Jiang Y (2022) Ultrasensitive photoelectrochemical biosensor based on black/red phosphorus heterojunction@Bi2Te3 hybrid and enzymatic signal amplification for the detection of colorectal cancer-related PiRNA-823. Sens Actuators B: Chem 368:132244. https://doi.org/10.1016/j.snb.2022.132244
Sytnikova YA, Rahman R, Chirn G-W, Clark JP, Lau NC (2014) Transposable element dynamics and PIWI regulation impacts LncRNA and gene expression diversity in Drosophila ovarian cell cultures. Genome Res 24(12):1977–1990. https://doi.org/10.1101/gr.178129.114
Tan Y, Liu L, Liao M, Zhang C, Hu S, Zou M, Gu M, Li X (2015) Emerging roles for PIWI proteins in cancer. Acta Biochim Biophys Sin 47(5):315–324. https://doi.org/10.1093/abbs/gmv018
Tan L, Mai D, Zhang B, Jiang X, Zhang J, Bai R, Ye Y, Li M, Pan L, Jiachun Su, Zheng Y, Liu Z, Zuo Z, Zhao Qi, Li X, Huang X, Yang J, Tan W, Zheng J, Lin D (2019) PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol Cancer 18(1):9. https://doi.org/10.1186/s12943-019-0940-3
Tan Y, Qin JN, Wan HQ, Zhao SM, Zhang C, Zeng Q, Qu SL (2022) PIWI/PiRNA-mediated regulation of signaling pathways in cell apoptosis. Eur Rev Med Pharmacol Sci 26(16)
Tan Y, Zeng Q, Sun X, Han Y, Zhao S, Wan H, Liu S, Zhang C, Fan W, Qu S (2024) PiRNA-823 exerts protective effects on high glucose concentration-induced HUVEC pyroptosis by targeting LCN2. https://doi.org/10.21203/rs.3.rs-4371914/v1
Tang X, Xie X, Wang X, Wang Y, Jiang X, Jiang H (2018) The combination of PiR-823 and eukaryotic initiation factor 3 B (EIF3B) activates hepatic stellate cells via upregulating TGF-Β1 in liver fibrogenesis. Med Sci Monit 24:9151–65. https://doi.org/10.12659/MSM.914222
Teleanu RI, Chircov C, Grumezescu AM, Teleanu DM (2019) Tumor angiogenesis and anti-angiogenic strategies for cancer treatment. J Clin Med 9(1):84. https://doi.org/10.3390/jcm9010084
Tong Y, Guan B, Sun Z, Dong X, Chen Y, Li Y, Jiang Y, Li J (2023) Ratiometric fluorescent detection of exosomal PiRNA-823 based on Au NCs/UiO-66-NH2 and target-triggered rolling circle amplification. Talanta 257:124307. https://doi.org/10.1016/j.talanta.2023.124307
Vourekas A, Alexiou P, Vrettos N, Maragkakis M, Mourelatos Z (2016) Sequence-dependent but not sequence-specific PiRNA adhesion traps MRNAs to the germ plasm. Nature 531(7594):390–394. https://doi.org/10.1038/nature17150
Vychytilova-Faltejskova P, Stitkovcova K, Radova L, Sachlova M, Kosarova Z, Slaba K, Kala Z, Svoboda M, Kiss I, Vyzula R, Cho WC, Slaby O (2018) Circulating PIWI-interacting RNAs PiR-5937 and PiR-28876 are promising diagnostic biomarkers of colon cancer. Cancer Epidemiol Biomark Prev 27(9):1019–1028. https://doi.org/10.1158/1055-9965.EPI-18-0318
Wang C, Lin H (2021) Roles of PiRNAs in transposon and pseudogene regulation of germline MRNAs and LncRNAs. Genome Biol 22(1):27. https://doi.org/10.1186/s13059-020-02221-x
Wang J, Shi Y, Zhou H, Zhang P, Song T, Ying Z, Yu H, Li Y, Zhao Yi, Zeng X, He S, Chen R (2022a) PiRBase: integrating PiRNA annotation in all aspects. Nucleic Acids Res 50(D1):D265–D272. https://doi.org/10.1093/nar/gkab1012
Wang S, Jiang X, Xie X, Yin J, Zhang J, Liu T, Chen S, Wang Y, Zhou X, Wang Y, Cui R, Jiang H (2022b) PiR-823 inhibits cell apoptosis via modulating mitophagy by binding to PINK1 in colorectal cancer. Cell Death Dis 13(5):465. https://doi.org/10.1038/s41419-022-04922-6
Wang H, Shi B, Zhang X, Shen P, He Q, Yin M, Pan Y, Ma J (2023) Exosomal Hsa-PiR1089 promotes proliferation and migration in neuroblastoma via targeting KEAP1. Pathol - Res Pract 241:154240. https://doi.org/10.1016/j.prp.2022.154240
Watanabe T, Cheng E-C, Zhong M, Lin H (2015) Retrotransposons and pseudogenes regulate MRNAs and LncRNAs via the PiRNA pathway in the germline. Genome Res 25(3):368–380. https://doi.org/10.1101/gr.180802.114
Weng W, Liu N, Toiyama Y, Kusunoki M, Nagasaka T, Fujiwara T, Wei Q, Qin H, Lin H, Ma Y, Goel A (2018) Novel evidence for a PIWI-interacting RNA (PiRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol Cancer 17(1):16. https://doi.org/10.1186/s12943-018-0767-3
Wu W-S, Huang W-C, Brown JS, Zhang D, Song X, Chen H, Tu S, Weng Z, Lee H-C (2018) PirScan: a webserver to predict PiRNA targeting sites and to avoid transgene silencing in C. Elegans. Nucleic Acids Res 46(W1):W43-48. https://doi.org/10.1093/nar/gky277
Wu W-S, Brown JS, Chen T-T, Chu Y-H, Huang W-C, Tu S, Lee H-C (2019) PiRTarBase: a database of PiRNA targeting sites and their roles in gene regulation. Nucleic Acids Res 47(D1):D181–D187. https://doi.org/10.1093/nar/gky956
Xiao B, Ma L, Merlin D (2017) Nanoparticle-mediated co-delivery of chemotherapeutic agent and SiRNA for combination cancer therapy. Expert Opin Drug Deliv 14(1):65–73. https://doi.org/10.1080/17425247.2016.1205583
Xin J, Du M, Jiang X, Wu Y, Ben S, Zheng R, Chu H, Li S, Zhang Z, Wang M (2021) Systematic evaluation of the effects of genetic variants on PIWI-interacting RNA expression across 33 cancer types. Nucleic Acids Res 49(1):90–97. https://doi.org/10.1093/nar/gkaa1190
Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E, Couté Y, Conn S, Kadlec J, Sachidanandam R, Kaksonen M, Cusack S, Ephrussi A, Pillai RS (2014) RNA clamping by Vasa assembles a PiRNA amplifier complex on transposon transcripts. Cell 157(7):1698–1711. https://doi.org/10.1016/j.cell.2014.05.018
Xu M, You Y, Hunsicker P, Hori T, Small C, Griswold MD, Hecht NB (2008) Mice deficient for a small cluster of Piwi-Interacting RNAs implicate piwi-interacting RNAs in transposon control. Biol Reprod 79(1):51–57. https://doi.org/10.1095/biolreprod.108.068072
Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang L, Chen L, Chu ZB, Tang B, Wang K, Wu XF, Xu J, Hu Y (2015) PiRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia 29(1):196–206. https://doi.org/10.1038/leu.2014.135
Yang L, Chen D, Duan R, Xia L, Wang J, Qurashi A, Jin P, Chen D (2007) Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development 134(23):4265–4272. https://doi.org/10.1242/dev.009159
Yao J, Xie M, Ma X, Song J, Wang Y, Xue X (2022) PIWI-interacting RNAs in cancer: biogenesis, function, and clinical significance. Front Oncol 12. https://doi.org/10.3389/fonc.2022.965684
Ye R (2016) ‘Pirnas variants and lung cancer risk: a post-Gwas study’. Public Health Theses 1335
Yin J, Jiang X-Y, Qi W, Ji C-G, Xie X-L, Zhang D-X, Cui Z-J, Wang C-K, Bai Y, Wang J, Jiang H-Q (2017) PiR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of <scp>HSF</Scp> 1. Cancer Sci 108(9):1746–1756. https://doi.org/10.1111/cas.13300
Yuan C, Qin H, Ponnusamy M, Chen Y, Lin Z (2021) PIWI-interacting RNA in cancer: molecular mechanisms and possible clinical implications (Review). Oncol Rep 46(3):209. https://doi.org/10.3892/or.2021.8160
Zhang W, Liu H, Yin J, Wu W, Zhu D, Amos CI, Fang S, Lee JE, Li Y, Han J, Wei Q (2016) Genetic variants in the PIWI-PiRNA pathway gene DCP1A predict melanoma disease-specific survival. Int J Cancer 139(12):2730–2737. https://doi.org/10.1002/ijc.30409
Zhang W, Yao G, Wang J, Yang M, Wang J, Zhang H, Li W (2020) NcRPheno: a comprehensive database platform for identification and validation of disease related noncoding RNAs. RNA Biol 17(7):943–955. https://doi.org/10.1080/15476286.2020.1737441
Zhang W, Zeng B, Yang M, Yang H, Wang J, Deng Y, Zhang H, Yao G, Wu S, Li W (2021) NcRNAVar: a manually curated database for identification of noncoding RNA variants associated with human diseases. J Mol Biol 433(11):166727. https://doi.org/10.1016/j.jmb.2020.166727
Zhang W, Zheng Z, Wang K, Mao W, Li X, Wang G, Zhang Y, Huang J, Zhang N, Wu P, Liu J, Zhang H, Che J, Peng B, Zheng J, Li W, Yao X (2023) PiRNA-1742 promotes renal cell carcinoma malignancy by regulating USP8 stability through binding to HnRNPU and thereby inhibiting MUC12 ubiquitination. Exp Mol Med 55(6):1258–1271. https://doi.org/10.1038/s12276-023-01010-3
Zhang H, Ali A, Gao J, Ban R, Jiang X, Zhang Y, Shi Q (2018) IsopiRBank: a research resource for tracking PiRNA isoforms. Database 2018. https://doi.org/10.1093/database/bay059
Zhao Q, Qian L, Guo Y, Lü J, Li D, Xie H, Wang Q, Ma W, Liu P, Liu Y, Wang T, Wu X, Han J, Yu Z (2023) IL11 signaling mediates PiR-2158 suppression of cell stemness and angiogenesis in breast cancer. Theranostics 13(7):2337–49. https://doi.org/10.7150/thno.82538
Zhu L, Ge J, Li T, Shen Y, Guo J (2019) TRNA-derived fragments and TRNA halves: the new players in cancers Zhu, L., Ge, J., Li, T., Shen, Y., & Guo, J. (2019). TRNA-derived fragments and TRNA halves: the new players in cancers. Cancer Letters, 452, 31–37. Https://Doi.Org/10.1016/j.Canlet.2019.03. Cancer Lett 452:31–37. https://doi.org/10.1016/j.canlet.2019.03.012
Zuo Y, Liang Y, Zhang J, Hao Y, Li M, Wen Z, Zhao Y (2019) Transcriptome analysis identifies Piwi-interacting RNAs as prognostic markers for recurrence of prostate cancer. Front Genet 10. https://doi.org/10.3389/fgene.2019.01018
Acknowledgements
The authors would like to appreciate the professional service of BioRender.com.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
All authors contributed equally for conceptualization, writing the draft and editing, visualization, and validation. Hamdy NM contributed software and in silico analysis. The authors confirm that no paper mill and artificial intelligence was used.
Corresponding author
Ethics declarations
Ethics approval
This is a review article. Ethical approval is not required for the study.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shaker, F.H., Sanad, E.F., Elghazaly, H. et al. piR-823 tale as emerging cancer-hallmark molecular marker in different cancer types: a step-toward ncRNA-precision. Naunyn-Schmiedeberg's Arch Pharmacol (2024). https://doi.org/10.1007/s00210-024-03308-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00210-024-03308-z