Abstract
Key message
The QTL hotspots determining seed glucosinolate content instead of only four HAG1 loci and elucidation of a potential regulatory model for rapeseed SGC variation.
Abstract
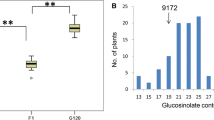
Glucosinolates (GSLs) are amino acid-derived, sulfur-rich secondary metabolites that function as biopesticides and flavor compounds, but the high seed glucosinolate content (SGC) reduces seed quality for rapeseed meal. To dissect the genetic mechanism and further reduce SGC in rapeseed, QTL mapping was performed using an updated high-density genetic map based on a doubled haploid (DH) population derived from two parents that showed significant differences in SGC. In 15 environments, a total of 162 significant QTLs were identified for SGC and then integrated into 59 consensus QTLs, of which 32 were novel QTLs. Four QTL hotspot regions (QTL-HRs) for SGC variation were discovered on chromosomes A09, C02, C07 and C09, including seven major QTLs that have previously been reported and four novel major QTLs in addition to HAG1 loci. SGC was largely determined by superimposition of advantage allele in the four QTL-HRs. Important candidate genes directly related to GSL pathways were identified underlying the four QTL-HRs, including BnaC09.MYB28, BnaA09.APK1, BnaC09.SUR1 and BnaC02.GTR2a. Related differentially expressed candidates identified in the minor but environment stable QTLs indicated that sulfur assimilation plays an important rather than dominant role in SGC variation. A potential regulatory model for rapeseed SGC variation constructed by combining candidate GSL gene identification and differentially expressed gene analysis based on RNA-seq contributed to a better understanding of the GSL accumulation mechanism. This study provides insights to further understand the genetic regulatory mechanism of GSLs, as well as the potential loci and a new route to further diminish the SGC in rapeseed.





Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Aarabi F, Kusajima M, Tohge T, Konishi T, Gigolashvili T, Takamune M, Sasazaki Y, Watanabe M, Nakashita H, Fernie AR, Saito K, Takahashi H, Hubberten HM, Hoefgen R, Maruyama-Nakashita A (2016) Sulfur deficiency-induced repressor proteins optimize glucosinolate biosynthesis in plants. Sci Adv 2:e1601087
Arcade A, Labourdette A, Falque M, Mangin B, Chardon F, Charcosset A, Joets J (2004) BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20:2324–2326
Bao BH, Chao HB, Wang H, Zhao WG, Zhang LN, Raboanatahiry N, Wang XD, Wang BS, Jia HB, Li MT (2018) Stable, environmental specific and novel QTL identification as well as genetic dissection of fatty acid metabolism in Brassica napus. Front Plant Sci 9:1018
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Cai G, Yang Q, Yi B, Fan C, Edwards D, Batley J, Zhou Y (2014) A complex recombination pattern in the genome of allotetraploid Brassica napus as revealed by a high-density genetic map. PLoS ONE 9:e109910
Cai G, Yang Q, Chen H, Yang Q, Zhang C, Fan C, Zhou Y (2016) Genetic dissection of plant architecture and yield-related traits in Brassica napus. Sci Rep 6:21625
Celenza JL, Quiel JA, Smolen GA, Merrikh H, Silvestro AR, Normanly J, Bender J (2005) The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol 137:253–262
Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, Correa M, Da Silva C, Just J, Falentin C, Koh CS, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao M, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier MC, Fan G, Renault V, Bayer PE, Golicz AA, Manoli S, Lee TH, Thi VH, Chalabi S, Hu Q, Fan C, Tollenaere R, Lu Y, Battail C, Shen J, Sidebottom CH, Wang X, Canaguier A, Chauveau A, Berard A, Deniot G, Guan M, Liu Z, Sun F, Lim YP, Lyons E, Town CD, Bancroft I, Wang X, Meng J, Ma J, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury JM, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou Y, Hua W, Sharpe AG, Paterson AH, Guan C, Wincker P (2014) Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345:950–953
Chao H, Wang H, Wang X, Guo L, Gu J, Zhao W, Li B, Chen D, Raboanatahiry N, Li M (2017) Genetic dissection of seed oil and protein content and identification of networks associated with oil content in Brassica napus. Sci Rep 7:46295
Chao H, Raboanatahiry N, Wang X, Zhao W, Chen L, Guo L, Li B, Hou D, Pu S, Zhang L, Wang H, Wang B, Li M (2019) Genetic dissection of harvest index and related traits through genome-wide quantitative trait locus mapping in Brassica napus L. Breed Sci 69:104–116
Chen S, Andreasson E (2001) Update on glucosinolate metabolism and transport. Plant Physiol Biochem 39:743–758
Dimov Z, Suprianto E, Hermann F, Mollers C (2012) Genetic variation for seed hull and fibre content in a collection of European winter oilseed rape material (Brassica napus L.) and development of NIRS calibrations. Plant Breeding 131:361–368
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
Ellerbrock B, Kim JH, Jander G (2007) Contribution of glucosinolate transport to Arabidopsis defense responses. Plant Signal Behav 2:282–283
Fahey JW, Zalcmann AT, Talalay P (2001) The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56:5–51
Falk KL, Tokuhisa JG, Gershenzon J (2007) The effect of sulfur nutrition on plant glucosinolate content: physiology and molecular mechanisms. Plant Biol (Stuttg) 9:573–581
Farnham MW, Wilson PE, Stephenson KK, Fahey JW (2004) Genetic and environmental effects on glucosinolate content and chemoprotective potency of broccoli. Plant Breed 123:60–65
Feng J, Long Y, Shi L, Shi JQ, Barker G, Meng JL (2012) Characterization of metabolite quantitative trait loci and metabolic networks that control glucosinolate concentration in the seeds and leaves of Brassica napus. New Phytol 193:96–108
Francisco M, Cartea ME, Butron AM, Sotelo T, Velasco P (2012) Environmental and genetic effects on yield and secondary metabolite production in Brassica rapa crops. J Agr Food Chem 60:5507–5514
Frerigmann H, Gigolashvili T (2014) MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol Plant 7:814–828
Frerigmann H, Pislewska-Bednarek M, Sanchez-Vallet A, Molina A, Glawischnig E, Gigolashvili T, Bednarek P (2016) Regulation of pathogen-triggered tryptophan metabolism in Arabidopsis thaliana by MYB transcription factors and indole glucosinolate conversion products. Mol Plant 9:682–695
Fu Y, Lu K, Qian LW, Mei JQ, Wei DY, Peng XH, Xu XF, Li JN, Frauen M, Dreyer F, Snowdon RJ, Qian W (2015) Development of genic cleavage markers in association with seed glucosinolate content in canola. Theor Appl Genet 128:1029–1037
Gigolashvili T, Berger B, Mock HP, Muller C, Weisshaar B, Fluegge UI (2007a) The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 50:886–901
Gigolashvili T, Yatusevich R, Berger B, Muller C, Flugge UI (2007b) The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 51:247–261
Graham N, May S (2011) Genetics and genomics of the Brassicaceae. Genetics and Genomics of the Brassicaceae
Grubb CD, Abel S (2006) Glucosinolate metabolism and its control. Trends Plant Sci 11:89–100
Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57:303–333
Hallauer AR, Filho J (1988) Quantitative genetics in maize breeding. Ames, Iowa State University Press, vol 10, p 468
Harper AL, Trick M, Higgins J, Fraser F, Clissold L, Wells R, Hattori C, Werner P, Bancroft I (2012) Associative transcriptomics of traits in the polyploid crop species Brassica napus. Nat Biotechnol 30:798–802
Harun S, Abdullah-Zawawi MR, Goh HH, Mohamed-Hussein ZA (2020) A comprehensive gene inventory for glucosinolate biosynthetic pathway in Arabidopsis thaliana. J Agric Food Chem 68:7281–7297
Hasan M, Friedt W, Pons-Kuehnemann J, Freitag NM, Link K, Snowdon RJ (2008) Association of gene-linked SSR markers to seed glucosinolate content in oilseed rape (Brassica napus ssp. napus). Theor Appl Genet 116:1035–1049
He Y, Fu Y, Hu D, Wei D, Qian W (2018) QTL mapping of seed glucosinolate content responsible for environment in Brassica napus. Front Plant Sci 9:891
Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, Goda H, Nishizawa OI, Shibata D, Saito K (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. P Natl Acad Sci USA 104:6478–6483
Howell PM, Sharpe AG, Lydiate DJ (2003) Homoeologous loci control the accumulation of seed glucosinolates in oilseed rape (Brassica napus). Genome 46:454–460
Hull AK, Vij R, Celenza JL (2000) Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci U S A 97:2379–2384
Huseby S, Koprivova A, Lee BR, Saha S, Mithen R, Wold AB, Bengtsson GB, Kopriva S (2013) Diurnal and light regulation of sulphur assimilation and glucosinolate biosynthesis in Arabidopsis. J Exp Bot 64:1039–1048
Jeschke V, Weber K, Moore SS, Burow M (2019) Coordination of glucosinolate biosynthesis and turnover under different nutrient conditions. Front Plant Sci 10:1560
Kondra ZP, Stefansson BR (1970) Inheritance of the major glucosinolates of rapeseed (Brassica napus) meal. Canjplant 50:643–647
Li HH, Ye GY, Wang JK (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374
Li HH, Ribaut JM, Li ZL, Wang JK (2008) Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor Appl Genet 116:243–260
Li F, Chen B, Xu K, Wu J, Song W, Bancroft I, Harper AL, Trick M, Liu S, Gao G, Wang N, Yan G, Qiao J, Li J, Li H, Xiao X, Zhang T, Wu X (2014) Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.). DNA Res 21:355–367
Li B, Zhao W, Li D, Chao H, Zhao X, Ta N, Li Y, Guan Z, Guo L, Zhang L, Li S, Wang H, Li M (2018) Genetic dissection of the mechanism of flowering time based on an environmentally stable and specific QTL in Brassica napus. Plant Sci 277:296–310
Li S, Zhu Y, Varshney RK, Zhan J, Zheng X, Shi J, Wang X, Liu G, Wang H (2020) A systematic dissection of the mechanisms underlying the natural variation of silique number in rapeseed (Brassica napus L.) germplasm. Plant Biotechnol J 18:568–580
Liu LZ, Qu CM, Wittkop B, Yi B, Xiao Y, He YJ, Snowdon RJ, Li JN (2013) A high-density SNP map for accurate mapping of seed fibre QTL in Brassica napus L. PLoS ONE 8:e83052
Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA, Zhao M, Ma J, Yu J, Huang S, Wang X, Wang J, Lu K, Fang Z, Bancroft I, Yang TJ, Hu Q, Wang X, Yue Z, Li H, Yang L, Wu J, Zhou Q, Wang W, King GJ, Pires JC, Lu C, Wu Z, Sampath P, Wang Z, Guo H, Pan S, Yang L, Min J, Zhang D, Jin D, Li W, Belcram H, Tu J, Guan M, Qi C, Du D, Li J, Jiang L, Batley J, Sharpe AG, Park BS, Ruperao P, Cheng F, Waminal NE, Huang Y, Dong C, Wang L, Li J, Hu Z, Zhuang M, Huang Y, Huang J, Shi J, Mei D, Liu J, Lee TH, Wang J, Jin H, Li Z, Li X, Zhang J, Xiao L, Zhou Y, Liu Z, Liu X, Qin R, Tang X, Liu W, Wang Y, Zhang Y, Lee J, Kim HH, Denoeud F, Xu X, Liang X, Hua W, Wang X, Wang J, Chalhoub B, Paterson AH (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 5:3930
Liu S, Huang H, Yi X, Zhang Y, Yang Q, Zhang C, Fan C, Zhou Y (2020) Dissection of genetic architecture for glucosinolate accumulations in leaves and seeds of Brassica napus by genome-wide association study. Plant Biotechnol J 18:1472–1484
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):1–21
Lu G, Harper AL, Trick M, Morgan C, Fraser F, O’Neill C, Bancroft I (2014) Associative transcriptomics study dissects the genetic architecture of seed glucosinolate content in Brassica napus. DNA Res 21:613–625
Lu K, Peng L, Zhang C, Lu J, Yang B, Xiao Z, Liang Y, Xu X, Qu C, Zhang K, Liu L, Zhu Q, Fu M, Yuan X, Li J (2017) Genome-wide association and transcriptome analyses reveal candidate genes underlying yield-determining traits in Brassica napus. Front Plant Sci 8:206
Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H (2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18:3235–3251
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations–science direct. Crop J 3:269–283
Miao L, Chao H, Chen L, Wang H, Zhao W, Li B, Zhang L, Li H, Wang B, Li M (2019) Stable and novel QTL identification and new insights into the genetic networks affecting seed fiber traits in Brassica napus. Theor Appl Genet 132:1761–1775
Nour-Eldin HH, Andersen TG, Burow M, Madsen SR, Jorgensen ME, Olsen CE, Dreyer I, Hedrich R, Geiger D, Halkier BA (2012) NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488:531–534
Olson-Manning CF, Lee CR, Rausher MD, Mitchell-Olds T (2013) Evolution of flux control in the glucosinolate pathway in Arabidopsis thaliana. Mol Biol Evol 30:14–23
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11:1650–1667
Qu CM, Li SM, Duan XJ, Fan JH, Jia LD, Zhao HY, Lu K, Li JN, Xu XF, Wang R (2015) Identification of candidate genes for seed glucosinolate content using association mapping in Brassica napus L. Genes (basel) 6:1215–1229
Raboanatahiry N, Chao H, Guo L, Gan J, Xiang J, Yan M, Zhang L, Yu L, Li M (2017) Synteny analysis of genes and distribution of loci controlling oil content and fatty acid profile based on QTL alignment map in Brassica napus. BMC Genomics 18:776
Rotmistrovsky K, Jang W, Schuler GD (2004) A web server for performing electronic PCR. Nucleic Acids Res 32:W108-112
Sharpe AG, Lydiate DJ (2003) Mapping the mosaic of ancestral genotypes in a cultivar of oilseed rape (Brassica napus) selected via pedigree breeding. Genome 46:461–468
Shen YS, Yang Y, Xu ES, Ge XH, Xiang Y, Li ZY (2018) Novel and major QTL for branch angle detected by using DH population from an exotic introgression in rapeseed (Brassica napus L.). Theor Appl Genet 131:67–78
Skirycz A, Reichelt M, Burow M, Birkemeyer C, Rolcik J, Kopka J, Zanor MI, Gershenzon J, Strnad M, Szopa J, Mueller-Roeber B, Witt I (2006) DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J 47:10–24
Sonderby IE, Geu-Flores F, Halkier BA (2010) Biosynthesis of glucosinolates - gene discovery and beyond. Trends Plant Sci 15:283–290
Song JM, Liu DX, Xie WZ, Yang Z, Guo L, Liu K, Yang QY, Chen LL (2021) BnPIR: Brassica napus pan-genome information resource for 1689 accessions. Plant Biotechnol J 19:412–414
Sun FM, Fan GY, Hu Q, Zhou YM, Guan M, Tong CB, Li JN, Du DZ, Qi CK, Jiang LC, Liu WQ, Huang SM, Chen WB, Yu JY, Mei DS, Meng JL, Zeng P, Shi JQ, Liu KD, Wang X, Wang XF, Long Y, Liang XM, Hu ZY, Huang GD, Dong CH, Zhang H, Li J, Zhang YL, Li LW, Shi CC, Wang JH, Lee SMY, Guan CY, Xu X, Liu SY, Liu X, Chalhoub B, Hua W, Wang HZ (2017) The high-quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. Plant J 92:452–468
Toroser D, Thormann CE, Osborn TC, Mithen R (1995) RFLP mapping of quantitative trait loci controlling seed aliphatic-glucosinolate content in oilseed rape (Brassica napus L). Theor Appl Genet 91:802–808
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578
Van Ooijen JW (2006) JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, Netherlands
Walker KC, Booth EJ (2001) Agricultural aspects of rape and other Brassica products. Eur J Lipid Sci Tech 103:441–446
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG, Park BS, Weisshaar B, Liu B, Li B, Liu B, Tong C, Song C, Duran C, Peng C, Geng C, Koh C, Lin C, Edwards D, Mu D, Shen D, Soumpourou E, Li F, Fraser F, Conant G, Lassalle G, King GJ, Bonnema G, Tang H, Wang H, Belcram H, Zhou H, Hirakawa H, Abe H, Guo H, Wang H, Jin H, Parkin IA, Batley J, Kim JS, Just J, Li J, Xu J, Deng J, Kim JA, Li J, Yu J, Meng J, Wang J, Min J, Poulain J, Wang J, Hatakeyama K, Wu K, Wang L, Fang L, Trick M, Links MG, Zhao M, Jin M, Ramchiary N, Drou N, Berkman PJ, Cai Q, Huang Q, Li R, Tabata S, Cheng S, Zhang S, Zhang S, Huang S, Sato S, Sun S, Kwon SJ, Choi SR, Lee TH, Fan W, Zhao X, Tan X, Xu X, Wang Y, Qiu Y, Yin Y, Li Y, Du Y, Liao Y, Lim Y, Narusaka Y, Wang Y, Wang Z, Li Z, Wang Z, Xiong Z, Zhang Z (2011) Brassica rapa genome sequencing project c the genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Wang X, Wang H, Long Y, Li D, Yin Y, Tian J, Chen L, Liu L, Zhao W, Zhao Y, Yu L, Li M (2013) Identification of QTLs associated with oil content in a high-oil Brassica napus cultivar and construction of a high-density consensus map for QTLs comparison in B. napus. PLoS One 8:e80569
Wang X, Yu K, Li H, Peng Q, Chen F, Zhang W, Chen S, Hu M, Zhang J (2015) High-density SNP map construction and QTL identification for the apetalous character in Brassica napus L. Front Plant Sci 6:1164
Wang XD, Chen L, Wang AN, Wang H, Tian JH, Zhao XP, Chao HB, Zhao YJ, Zhao WG, Xiang J, Gan JP, Li MT (2016) Quantitative trait loci analysis and genome-wide comparison for silique related traits in Brassica napus. Bmc Plant Biol 16(1):1–5
Wang B, Wu ZK, Li ZH, Zhang QH, Hu JL, Xiao YJ, Cai DF, Wu JS, King GJ, Li HT, Liu KD (2018) Dissection of the genetic architecture of three seed-quality traits and consequences for breeding in Brassica napus. Plant Biotechnol J 16:1336–1348
Wang S, Basten C, Zeng Z (2012) Windows QTL Cartographer v2.5. Department of Statistics. North Carolina State University, Raleigh, NC
Wei L, Jian H, Lu K, Filardo F, Yin N, Liu L, Qu C, Li W, Du H, Li J (2016) Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol J 14:1368–1380
Wei D, Cui Y, Mei J, Qian L, Lu K, Wang ZM, Li J, Tang Q, Qian W (2019) Genome-wide identification of loci affecting seed glucosinolate contents in Brassica napus L. J Integr Plant Biol 61:611–623
Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7:263–270
Yan X, Chen S (2007) Regulation of plant glucosinolate metabolism. Planta 226:1343–1352
Ye J, Yang Y, Chen B, Shi J, Luo M, Zhan J, Wang X, Liu G, Wang H (2017) An integrated analysis of QTL mapping and RNA sequencing provides further insights and promising candidates for pod number variation in rapeseed (Brassica napus L.). BMC Genomics 18:71
Zhao J, Meng J (2003) Detection of loci controlling seed glucosinolate content and their association with Sclerotinia resistance in Brassica napus. Plant Breed 122:19–23
Zhao WG, Wang XD, Wang H, Tian JH, Li BJ, Chen L, Chao HB, Long Y, Xiang J, Gan JP, Liang WS, Li MT (2016) Genome-wide identification of QTL for seed yield and yield-related traits and construction of a high-density consensus map for QTL comparison in Brassica napus. Front Plant Sci 7:17
Zhao WG, Zhang LN, Chao HB, Wang H, Ta N, Li HX, Wang XD, Li SS, Xiang J, Zhang LB, Li MT (2019) Genome-wide identification of silique-related traits based on high-density genetic linkage map in Brassica napus. Mol Breeding 39:86
Acknowledgements
This work was financially supported by National Key R&D Program of China (2016YFD0101300) and National Natural Science Foundation of China (31871656 and 32001583).
Author information
Authors and Affiliations
Contributions
HC carried out QTL analysis, transcriptome analysis and wrote the manuscript. HL and SY participated in the field experiment and collected data. WZ and KC performed the data processing, detection and modifying of pictures. HW and NR provided many guidelines for the paper and revised the manuscript. JH and ML designed, led and coordinated the overall study.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that the experiments comply with the current laws of the country in which they were performed.
Additional information
Communicated by Isobel AP Parkin.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
122_2022_4161_MOESM1_ESM.tif
Supplementary file1 Fig. S1 Collinearity of the updated high-density genetic map and the previous high-density map Chao et al., (2017) constructed with the ‘Darmor-bzh’ reference genome. Outer and inner circles represent 19 genetic linkage groups of the genetic map and 19 chromosomes of the reference genome. The arrows with same color indicate the inferior collinearity regions in previous high-density map and its corresponding collinearity regions in the current updated high-density map when comparison to the reference genome. (TIF 8383 kb)
122_2022_4161_MOESM2_ESM.jpg
Fig. S2 QTL hotspot identified on chromosome A09 and candidate genes within the QTL hotspot. The identified QTLs for SGC in different environments are shown by curves above the line of the linkage group, and corresponding additive effects are display in curves of the same color below the linkage group. The short-terms localization in the middle represent identified QTL detected at the corresponding position, and subsequently are integrated into consensus QTL showed by brown term with yellow line near the peak, and the name of major QTL are displayed in red fonts. At the bottom, corresponding part of the reference genome ‘Darmor-bzh’ and ‘ZS11’, candidate gene within this region and collinearity are shown.
122_2022_4161_MOESM4_ESM.tif
Supplementary file4 Fig S4 Statistics of differentially expressed genes between Ken-C8 and N53-2 in leaf and seed. (TIF 107 kb)
122_2022_4161_MOESM7_ESM.tif
Supplementary file7 Fig. S7 The relative expression level of BnaC02. MYB34 and BnaC09. MYB34 (A) and seed indole GSLs content in the two parents (B). I3M: Indol-3-yl-methy-GSL; 4MTI3M: 4-methoxyindol-3-yl-methy-GSL; 1MTI3M: 1- methoxyindol-3-yl-methy-GSL. (TIF 225 kb)
122_2022_4161_MOESM8_ESM.jpg
Fig. S8 Verifying the gene expression difference from RNA-Seq analysis by qRT-PCR. The level of differential expression on the left side was calculated by FPKM value from RNA-Seq, and the right side by qRT-PCR.
Rights and permissions
About this article
Cite this article
Chao, H., Li, H., Yan, S. et al. Further insight into decreases in seed glucosinolate content based on QTL mapping and RNA-seq in Brassica napus L. Theor Appl Genet 135, 2969–2991 (2022). https://doi.org/10.1007/s00122-022-04161-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-022-04161-5