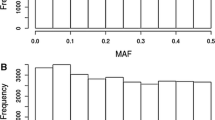
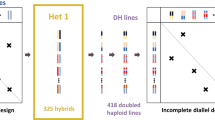
Abstract
Key message
Silage quality traits of maize hybrids between the Dent and Flint heterotic groups mostly involved QTL specific of each parental group, some of them showing unfavorable pleiotropic effects on yield.
Abstract
Maize (Zea mays L.) is commonly used as silage for cattle feeding in Northern Europe. In addition to biomass production, improving whole-plant digestibility is a major breeding objective. To identify loci involved in the general (GCA, parental values) and specific combining ability (SCA, cross-specific value) components of hybrid value, we analyzed an incomplete factorial design of 951 hybrids obtained by crossing inbred lines issued from two multiparental connected populations, each specific to one of the heterotic groups used for silage in Europe (“Dent” and “Flint”). Inbred lines were genotyped for approximately 20K single nucleotide polymorphisms, and hybrids were phenotyped in eight environments for seven silage quality traits measured by near-infrared spectroscopy, biomass yield and precocity (partly analyzed in a previous study). We estimated variance components for GCA and SCA and their interaction with environment. We performed QTL detection using different models adapted to this hybrid population. Strong family effects and a predominance of GCA components compared to SCA were found for all traits. In total, 230 QTL were detected, with only two showing SCA effects significant at the whole-genome level. More than 80% of GCA QTL were specific of one heterotic group. QTL explained individually less than 5% of the phenotypic variance. QTL co-localizations and correlation between QTL effects of quality and productivity traits suggest at least partial pleiotropic effects. This work opens new prospects for improving maize hybrid performances for both biomass productivity and quality accounting for complementarities between heterotic groups.





Similar content being viewed by others
References
Andrieu J (1995) Prévision de la digestibilité et de la valeur énergétique du maïs fourrage à l’état frais. INRA Prod Anim 8(4):273–274
Argillier O, Barrière Y, Hebert Y (1995) Genetic-variation and selection criterion for digestibility traits of forage maize. Euphytica 82(2):175–184
Argillier O, Mechin V, Barrière Y (2000) Inbred line evaluation and breeding for digestibility-related traits in forage maize. Crop Sci 40(6):1596–1600
Badji A, Otim M, Machida L, Odong T, Bomet Kwemoi D, Okii D, Agbahoungba S et al (2018) Maize combined insect resistance genomic regions and their co-localization with cell wall constituents revealed by tissue-specific QTL meta-analyses. Front Plant Sci 9:895
Baker CW, Givens DI, Deaville ER (1994) Prediction of organic matter digestibility in vivo of grass silage by near infrared reflectance spectroscopy: effect of calibration method, residual moisture and particle size. Anim Feed Sci Technol 50(1):17–26
Baldy A, Jacquemot MP, Griveau Y, Bauland C, Reymond M, Mechin V (2017) Energy values of registered corn forage hybrids in France over the last 20 years rose in a context of maintained yield increase. Am J Plant Sci 08(06):1449–1461
Barrière Y, Emile JC (2000) Le maïs fourrage: III. évaluation et perspectives de progrès génétiques sur les caractères de valeurs alimentaire. Fourrages 163:221–238
Barrière Y, Guillet C, Goffner D, Pichon M (2003) Genetic variation and breeding strategies for improved cell wall digestibility in annual forage crops—a review. Anim Res 52(3):193–228
Barrière Y, Riboulet C, Méchin V, Maltese S, Pichon M, Cardinal A, Lapierre C, Lubberstedt T, Martinant JP (2007) Genetics and genomics of lignification in grass cell walls based on maize as model species. Genes Genomes Genomics 1:133–156
Barrière Y, Thomas J, Denoue D (2008) QTL mapping for lignin content, lignin monomeric composition, p-hydroxycinnamate content, and cell wall digestibility in the maize recombinant inbred line progeny F838 × F286. Plant Sci 175(4):585–595
Barrière Y, Méchin V, Denoue D, Bauland C, Laborde J (2010) QTL for yield, earliness, and cell wall quality traits in topcross experiments of the F838 × F286 early maize RIL progeny. Crop Sci 50(5):1761–1772
Barrière Y, Courtial A, Chateigner-Boutin A-L, Denoue D, Grima-Pettenati J (2016) Breeding maize for silage and biofuel production, an illustration of a step forward with the genome sequence. Plant Sci 242(January):310–329
Barrière Y, Guillaumie S, Denoue D, Pichon M, Goffner D, Martinant JP (2017) Investigating the unusually high cell wall digestibility of the old INRA early flint F4 maize inbred line. Maydica 62:M31
Barros-Rios J, Malvar RA, Jung H-JG, Santiago R (2011) Cell wall composition as a maize defence mechanism against corn borers. Phytochemistry 72(4):365–371
Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81(5):1084–1097
Buendgen MR, Coors JG, Grombacher AW, Russell WA (1990) European corn borer resistance and cell wall composition of three maize populations. Crop Sci 30(3):505–510
Butler DG, Cullis BR, Gilmour AR, Gogel BJ (2007) ASReml-R reference manual. The State of Queensland, Department of Primary Industries and Fisheries, Brisbane
Cardinal AJ, Lee M (2005) Genetic relationships between resistance to stalk-tunneling by the european corn borer and cell wall components in maize population B73 × B52. Theor Appl Genet 111(1):1–7
Cardinal A, Lee M, Moore K (2003) Genetic mapping and analysis of quantitative trait loci affecting fiber and lignin content in maize. Theor Appl Genet 106(5):866–874
Cornu A, Besle JM, Mosoni P, Grenet E (1994) Lignin–carbohydrate complexes in forages: structure and consequences in the ruminal degradation of cell wall carbohydrates. Reprod Nutr Dev 34(5):385–398
Courtial A, Thomas J, Reymond M, Méchin V, Grima-Pettenati J, Barrière Y (2013) Targeted linkage map densification to improve cell wall related QTL detection and interpretation in maize. Theor Appl Genet 126(5):1151–1165
Courtial A, Méchin V, Reymond M, Grima-Pettenati J, Barrière Y (2014) Colocalizations between several QTLs for cell wall degradability and composition in the F288 × F271 early maize RIL progeny raise the question of the nature of the possible underlying determinants and breeding targets for biofuel capacity. Bioenergy Res 7(1):142–156
Ducrocq S, Giauffret C, Madur D, Combes V, Dumas F, Jouanne S, Coubriche D, Jamin P, Moreau L, Charcosset A (2009) Fine mapping and haplotype structure analysis of a major flowering time quantitative trait locus on maize chromosome 10. Genetics 183(4):1555–1563
El Hage F, Legland D, Borrega N, Jacquemot M-P, Griveau Y, Coursol S, Méchin V, Reymond M (2018) Tissue lignification, cell wall p-coumaroylation and degradability of maize stems depend on water status. J Agric Food Chem 66(19):4800–4808
Flint-Garcia SA, Jampatong C, Darrah LL, McMullen MD (2003) Quantitative Trait Locus analysis of stalk strength in four maize populations. Crop Sci 43(1):13
Fontaine AS, Bout S, Barrière Y, Vermerris W (2003) Variation in cell wall composition among forage maize (Zea Mays L.) inbred lines and its impact on digestibility: analysis of neutral detergent fiber composition by pyrolysis–gas chromatography–mass spectrometry. J Agric Food Chem 51(27):8080–8087
Gao X, Starmer J, Martin ER (2008) A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol 32(4):361–369
Giordano A, Liu Z, Panter SN, Dimech AM, Shang Y, Wijesinghe H, Fulgueras K et al (2014) Reduced lignin content and altered lignin composition in the warm season forage grass paspalum dilatatum by down-regulation of a cinnamoyl CoA reductase gene. Transgenic Res 23(3):503–517
Giraud H, Bauland C, Falque M, Madur D, Combes V, Jamin P, Monteil C et al (2017a) Linkage analysis and association mapping QTL detection models for hybrids between multiparental populations from two heterotic groups: application to biomass production in maize (Zea Mays L.). G3 7(11):3649–3657
Giraud H, Bauland C, Falque M, Madur D, Combes V, Jamin P, Monteil C et al (2017b) Reciprocal genetics: identifying QTL for general and specific combining abilities in hybrids between multiparental populations from two maize (Zea Mays L.) heterotic groups. Genetics 207(3):1167–1180
Goering HK, Van Soest PJ (1970) Forage fiber analyses (apparatus, reagents, procedures, and some applications). U.S. Agricultural Research Service Handbook, pp 1–379
Grabber JH, Quideau S, Ralph J (1996) P-coumaroylated syringyl units in maize lignin: implications for β-ether cleavage by thioacidolysis. Phytochemistry 43(6):1189–1194
Hartley RD (1972) p-Coumaric and ferulic acid components of cell walls of ryegrass and their relationships with lignin and digestibility. J Sci Food Agric 23(11):1347–1354
Hickey JM, Gorjanc G, Varshney RK, Nettelblad C (2015) Imputation of single nucleotide polymorphism genotypes in biparental, backcross, and topcross populations with a hidden markov model. Crop Sci 55(5):1934
Jung HG, Vogel KP (1986) Influence of lignin on digestibility of forage cell wall material. J Anim Sci 62(6):1703–1712
Jung HG, Mertens DR, Payne AJ (1997) Correlation of acid detergent lignin and Klason lignin with digestibility of forage dry matter and neutral detergent fiber. J Dairy Sci 80(8):1622–1628
Khan NA, Yu P, Ali M, Cone JW, Hendriks WH (2015) Nutritive value of maize silage in relation to dairy cow performance and milk quality. J Sci Food Agric 95(2):238–252
Krakowsky MD, Lee M, Beeghly HH, Coors JG (2003) Characterization of quantitative trait loci affecting fiber and lignin in maize (Zea Mays L.). Maydica 48(4):283–292
Krakowsky MD, Lee M, Coors JG (2005) Quantitative trait loci for cell wall components in recombinant inbred lines of maize (Zea Mays L.) I: stalk tissue. Theor Appl Genet 111(2):337–346
Krakowsky MD, Lee M, Coors JG (2006) Quantitative trait loci for cell wall components in recombinant inbred lines of maize (Zea Mays L.) II: leaf sheath tissue. Theor Appl Genet 112(4):717–726
Lawrence CJ, Dong Q, Polacco ML, Seigfried TE, Brendel V (2004) MaizeGDB, the community database for maize genetics and genomics. Nucleic Acids Res 32(Database issue):D393–D397
Le Gall H, Philippe F, Domon J-M, Gillet F, Pelloux J, Rayon C (2015) Cell wall metabolism in response to abiotic stress. Plants 4(1):112–166
Leng P, Ouzunova M, Landbeck M, Wenzel G, Eder J, Darnhofer B, Lübberstedt T (2018) Quantitative trait loci mapping of forage stover quality traits in six mapping populations derived from European elite maize germplasm. Plant Breed 137(2):139–147
Li K, Wang H, Hu X, Liu Z, Wu Y, Huang C (2016) Genome-wide association study reveals the genetic basis of stalk cell wall components in maize. PLoS ONE 11(8):e0158906
Lübberstedt T, Melchinger AE, Schön CC, Utz HF, Klein D (1997a) QTL mapping in testcrosses of European flint lines of maize: I. Comparison of different testers for forage yield traits. Crop Sci 37(3):921–931
Lübberstedt T, Melchinger AE, Klein D, Degenhardt H, Paul C (1997b) QTL mapping in testcrosses of European flint lines of maize: II. Comparison of different testers for forage quality traits. Crop Sci 37(6):1913–1922
Lübberstedt T, Melchinger AE, Fähr S, Klein D, Dally A, Westhoff P (1998) QTL mapping in testcrosses of flint lines of maize: III. Comparison across populations for forage traits. Crop Sci 38(5):1278–1289
Méchin V, Argillier O, Menanteau V, Barrière Y, Mila I, Pollet B, Lapierre C (2000) Relationship of cell wall composition to in vitro cell wall digestibility of maize inbred line stems. J Sci Food Agric 80(5):574–580
Méchin V, Argillier O, Hébert Y, Guingo E, Moreau L, Charcosset A, Barrière Y (2001) Genetic analysis and QTL mapping of cell wall digestibility and lignification in silage maize. Crop Sci 41(3):690–697
Méchin V, Argillier O, Rocher F, Hébert Y, Mila I, Pollet B, Barriére Y, Lapierre C (2005) In search of a maize ideotype for cell wall enzymatic degradability using histological and biochemical lignin characterization. J Agric Food Chem 53(15):5872–5881
Moore KJ, Jung H-JG (2001) Lignin and fiber digestion. J Range Manag 54(4):420–430
Nakagawa S, Schielzeth H (2013) A general and simple method for obtaining R² from generalized linear mixed-effects models. Methods Ecol Evol 4:133–142
Penning BW, Sykes RW, Babcock NC, Dugard CK, Held MA, Klimek JF, Shreve JT et al (2014) Genetic determinants for enzymatic digestion of lignocellulosic biomass are independent of those for lignin abundance in a maize recombinant inbred population1. Plant Physiol 165(4):1475–1487
R Core Team (2013) R: a language and environment for statistical computing. R foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org
Ralph J, Hatfield RD, Quideau S, Helm RF, Grabber JH, Jung H-JG (1994) Pathway of p-coumaric acid incorporation into maize lignin as revealed by NMR. J Am Chem Soc 116(21):9448–9456
Reif JC, Gumpert F-M, Fischer S, Melchinger AE (2007) Impact of interpopulation divergence on additive and dominance variance in hybrid populations. Genetics 176(3):1931–1934
Riboulet C, Fabre F, Dénoue D, Martinant JP, Lefevre B, Barrière Y (2008) QTL mapping and candidate gene research from lignin content and cell wall digestibility in a top-cross of a flint maize recombinant inbred line progeny harvested at silage stage. Maydica 53(1):1
Roussel V, Gibelin C, Fontaine A-S, Barrière Y (2002) Genetic analysis in recombinant inbred lines of early dent forage maize. II—QTL mapping for cell wall constituents and cell wall digestibility from per se value and top cross experiments. Maydica 47(January):9–20
Salvi S, Tuberosa R, Chiapparino E, Maccaferri M, Veillet S, van Beuningen L, Isaac P, Edwards K, Phillips RL (2002) Toward positional cloning of vgt1, a QTL controlling the transition from the vegetative to the reproductive phase in maize. Plant Mol Biol 48(5–6):601–613
Santiago R, Barros-Rios J, Malvar RA (2013) Impact of cell wall composition on maize resistance to pests and diseases. Int J Mol Sci 14(4):6960–6980
Sosnowski O, Charcosset A, Joets J (2012) BioMercator V3: an upgrade of genetic map compilation and quantitative trait loci meta-analysis algorithms. Bioinformatics 28(15):2082–2083
Stuth J, Jama A, Tolleson D (2003) Direct and indirect means of predicting forage quality through near infrared reflectance spectroscopy. Field Crops Res 84(1–2):45–56
Surault F, Emile JC, Briand M, Barrière Y, Traineau R (2005) Variabilité génétique de la digestibilité in vivo d’hybrides de maïs. Bilan de 34 années de mesures. Fourrages 183:459–474
Torres AF, Noordam-Boot CMM, Dolstra O, van der Weijde T, Combes E, Dufour P, Vlaswinkel L, Visser RGF, Trindade LM (2014) Cell wall diversity in forage maize: genetic complexity and bioenergy potential. Bioenergy Res 8(1):187–202
Torres AF, Noordam-Boot CMM, Dolstra O, Vlaswinkel L, Visser RGF, Trindade LM (2015) Extent of genotypic variation for maize cell wall bioconversion traits across environments and among hybrid combinations. Euphytica 206(2):501–511
Truntzler M, Barrière Y, Sawkins MC, Lespinasse D, Betran J, Charcosset A, Moreau L (2010) Meta-analysis of QTL involved in silage quality of maize and comparison with the position of candidate genes. Theor Appl Genet 121(8):1465–1482
Vermerris W, Sherman DM, McIntyre LM (2010) Phenotypic plasticity in cell walls of maize brown midrib mutants is limited by lignin composition. J Exp Bot 61(9):2479–2490
Wang H, Li K, Hu X, Liu Z, Wu Y, Huang C (2016) Genome-wide association analysis of forage quality in maize mature stalk. BMC Plant Biol 16:227
Williams E, Piepho H-P, Whitaker D (2011) Augmented p-rep designs. Biom J 53(1):19–27
Wolak ME (2012) Nadiv : an R package to create relatedness matrices for estimating non-additive genetic variances in animal models. Methods Ecol Evol 3(5):792–796
Zhang Y, Culhaoglu T, Pollet B, Melin C, Denoue D, Barrière Y, Baumberger S, Méchin V (2011) Impact of lignin structure and cell wall reticulation on maize cell wall degradability. J Agric Food Chem 59(18):10129–10135
Zhang Y, Yang D-Q, Wang X-M, Feng M, Brunette G (2012) Process for fungal modification of lignin and preparing wood adhesives with the modified lignin and wood composites made from such adhesives. Patent: WO 2012/113058 A1
Acknowledgements
A. I. Seye’s Ph.D. was funded by the Senegalese Institute of Agricultural Research (ISRA) through a Scholarship from the West Africa Agricultural Productivity Program (WAAPP) given by the National Institute of Higher Education in Agricultural Sciences—Montpellier SupAgro. We are grateful to Caussade Semences, Euralis Semences, Limagrain Europe, Maïsadour Semences, Pioneer Genetics, R2n, Syngenta Seeds and KWS grouped in the frame of the ProMais “SAM-MCR” program for the funding, inbred lines development, hybrid production and phenotyping. We also thank scientists from these companies for helpful discussions on the results. We thank Magali Joannin of the INRA station of GQE Le Moulon for seed management and D. Madur and V. Combes for DNA extraction and genotyping analyses. We thank C. Palaffre and the INRA experimental unit of Saint-Martin-de-Hinx for seed production and seed management. We thank Dominique Kermarrec and the INRA station of Ploudaniel for their support to ProMaïs in phenotyping. We thank Pierre Dardenne from CRA-Wallonie for the NIRS prediction equations of silage quality traits.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that the experiments comply with the current laws of the countries in which the experiments were performed.
Additional information
Communicated by Thomas Lubberstedt.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Seye, A.I., Bauland, C., Giraud, H. et al. Quantitative trait loci mapping in hybrids between Dent and Flint maize multiparental populations reveals group-specific QTL for silage quality traits with variable pleiotropic effects on yield. Theor Appl Genet 132, 1523–1542 (2019). https://doi.org/10.1007/s00122-019-03296-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03296-2