Abstract
Key message A single recessive gene for complete resistance to powdery mildew and a major-effect QTL for partial resistance to downy mildew were co-localized in a Cucumis hystrix introgression line of cucumber.
Abstract
Downy mildew (DM) and powdery mildew (PM) are two major foliar diseases in cucumber. DM resistance (DMR) and PM resistance (PMR) may share common components; however, the genetic relationship between them remains unclear. IL52, a Cucumis hystrix introgression line of cucumber which has been reported to possess DMR, was recently identified to exhibit PMR as well. In this study, a single recessive gene pm for PMR was mapped to an approximately 468-kb region on chromosome 5 with 155 recombinant inbred lines (RILs) and 193 F2 plants derived from the cross between a susceptible line ‘changchunmici’ and IL52. Interestingly, pm was co-localized with the major-effect DMR QTL dm5.2 confirmed by combining linkage analysis and BSA-seq, which was consistent with the observed linkage of DMR and PMR in IL52. Further, phenotype–genotype correlation analysis of DMR and PMR in the RILs indicated that the co-localized locus pm/dm5.2 confers complete resistance to PM and partial resistance to DM. Seven candidate genes for DMR were identified within dm5.2 by BSA-seq analysis, of which Csa5M622800.1, Csa5M622830.1 and Csa5M623490.1 were also the same candidate genes for PMR. A single nucleotide polymorphism that is present in the 3ˊ untranslated region (3′UTR) of Csa5M622830.1 co-segregated perfectly with PMR. The GATA transcriptional factor gene Csa5M622830.1 may be a likely candidate gene for DMR and PMR. This study has provided a clear evidence for the relationship between DMR and PMR in IL52 and sheds new light on the potential value of IL52 for cucumber DMR and PMR breeding program.






Similar content being viewed by others
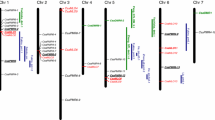
References
Abe A, Kosugi S, Yoshida K et al (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 119:313–327
An Y, Han X, Tang X et al (2014) Poplar GATA transcription factor PdGNC is capable of regulating chloroplast ultrastructure, photosynthesis, and vegetative growth in Arabidopsis under varying nitrogen levels. Plant Cell Tissue Organ Cult 30:174–178
Bai Z, Yuan X, Cai R et al (2008) QTL analysis of downy mildew resistance in cucumber. Prog Nat Sci 18:706–710 (in Chinese)
Berg JA, Appiano M, Martínez MS et al (2015) A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant Biol 15:243
Bi Y, Zhang Y, Signorelli T et al (2005) Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J 44:680–692
Caldwell D, Chan E, de Vries J et al (2011) Methods and compositions for identifying downy mildew resistant cucumber plants. United States patent US 2011/0126309 A1
Cao Q (2006) Research on cucumber alien translocation line possessing resistance to downy mildew and its application in cucumber breeding. Dissertation, Nanjing Agricultural University (in Chinese)
Cavagnaro PF, Senalik DA, Yang L et al (2010) Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genom 11:569
Chen J, Kirkbride J (2000) A new synthetic species Cucumis (Cucurbitaceae) from interspecific hybridization and chromosome doubling. Brittonia 52:315–319
Chen J, Staub J, Tashiro Y et al (1997) Successful interspecific hybridization between Cucumis sativus L. and C. hystrix Chakr. Euphytica 96:413–419
Chen J, Luo X, Staub J et al (2003) An allotriploid derived from a amphidiploid × diploid. Euphytica 131:235–241
Cockerham CC (1983) Covariances of relatives from self-fertilization. Crop Sci 23:1177–1180
Das S, Upadhyaya H, Bajaj D et al (2015) Deploying QTL-seq for rapid delineation of a potential candidate gene underlying major trait-associated QTL in chickpea. DNA Res 22:193–203
de Ruiter W, Hofstede R, de Vries J, van den Heuvel H (2008) Combining QTL for resistance to CYSDV and powdery mildew in a single cucumber line. In: Proceedings of 9th EUCARPIA meeting on genetics and breeding of Cucurbitaceae (Pitrat M, ed), INRA, Avignon (France), 21–24 May, pp 181–188
Dijkhuizen A, Kennard WC, Havey MJ et al (1996) RFLP variation and genetic relationships in cultivated cucumber. Euphytica 90:79–87
Ding G, Qin Z, Zhou X et al (2007) RAPD and SCAR markers linked to downy mildew resistance genes in cucumber. Acta Bot Boreali-Occident Sin 27:1747–1751 (in Chinese)
Epps W, Barnes W (1952) The increased susceptibility of the Palmetto cucumber to downy mildew in South Carolina. Plant Dis Rep 36:14–15
Evangelisti E, Rey T, Schornack S (2014) Cross-interference of plant development and plant-microbe interactions. Curr Opin Plant Biol 20:118–126
Fukino N, Yoshioka Y, Sugiyama M et al (2013) Identification and validation of powdery mildew (Podosphaera xanthii)-resistant loci in recombinant inbred lines of cucumber (Cucumis sativus L.). Mol Breed 32:267–277
Gao D, Appiano M, Huibers RP et al (2015) Natural loss-of-function mutation of EDR1 conferring resistance to tomato powdery mildew in Arabidopsis thaliana accession C24. Mol Plant Pathol 16:71–82
He X, Li Y, Pandey S et al (2013) QTL mapping of powdery mildew resistance in WI 2757 cucumber (Cucumis sativus L.). Theor Appl Genet 129:819–829
He H, Zhu S, Jiang Z et al (2016) Comparative mapping of powdery mildew resistance gene Pm21 and functional characterization of resistance-related genes in wheat. Theor Appl Genet 129:819–829
Horejsi T, Staub JE (1999) Genetic variation in cucumber (Cucumis sativus L.) as assessed by random amplified polymorphic DNA. Genet Resour Crop Evol 46:337–350
Huibers RP, Loonen AEHM, Gao D et al (2013) Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS ONE 8:e67467
Illa-Berenguer E, Van Houten J, Huang Z (2015) Rapid and reliable identification of tomato fruit weight and locule number loci by QTL-seq. Theor Appl Genet 128:1329–1342
Jenkins SF, Wehner TC (1983) A system for the measurement of foliar diseases of cucumber. Cucurbit Genet Coop Rep 6:10–12
Kooistra E (1968) Powdery mildew resistance in cucumber. Euphytica 17:236–244
Kozma P, Dula T (2003) Inheritance of resistance to downy mildew and powdery mildew of hybrid family Muscadinia × V. vinifera × V. amurensis × Franco-American hybrid. Acta Hortic 603:457–463
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760
Li Y, Yang L, Pathak M et al (2011) Fine genetic mapping of cp: a recessive gene for compact (dwarf) plant architecture in cucumber, Cucumis sativus L. Theor Appl Genet 123:973–983
Liu L, Yuan X, Cai R et al (2008) Quantitative trait loci for resistance to powdery mildew in cucumber under seedling spray inoculation and leaf disc infection. J Phytopathol 156:691–697
Lu H, Lin T, Klein J et al (2014) QTL-seq identifies an early flowering QTL located near flowering locus T in cucumber. Theor Appl Genet 127:1491–1499
McGrath MT (2001) Fungicide resistance in cucurbit powdery mildew: experiences and challenges. Plant Dis 85:236–245
Merdinoglu D, Wiedemann-Merdinoglu S, Coste P et al (2003) Genetic analysis of downy mildew resistance derived from Muscadinia rotundifolia. Acta Hortic 603:451–456
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Nie J, He H, Peng J et al (2015a) Identification and fine mapping of pm5.1: a recessive gene for powdery mildew resistance in cucumber (Cucumis sativus L.). Mol Breed 35:7
Nie J, Wang Y, He H et al (2015b) Loss-of-function mutations in CsMLO1 confer durable powdery mildew resistance in cucumber (Cucumis sativus L.). Front. Plant Sci 6:1155
Olaya G, Kuhn P, Hert A (2009) Fungicide resistance in cucurbit downy mildew. Phytopathology 9:S171
Olczak-Woltman H, Marcinkowska J, Niemirowicz-Szczytt K (2011) The genetic basis of resistance to downy mildew in Cucumis spp.—latest developments and prospects. J Appl Genet 52:249–255
Pandey M, Khan A, Singh V et al (2017) QTL-seq approach identified genomic regions and diagnostic markers for rust and late leaf spot resistance in groundnut (Arachis hypogaea L.). Plant Biotechnol J 15:927–941
Pang X, Zhou X, Wan H et al (2013) QTL mapping of downy mildew resistance in an introgression line derived from interspecific hybridization between cucumber and Cucumis hystrix. J Phytopathol 161:536–543
Perchepied L, Bardin M, Dogimont C, Pitrat M (2005) Relationship between loci conferring downy mildew and powdery mildew resistance in melon assessed by quantitative trait loci mapping. Phytopathology 95:556–565
Pérez-García A, Romero D, FernÁndez-OrtuÑo D et al (2009) The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits. Mol Plant Pathol 10:153–160
Ren Y, Zhang Z, Liu J et al (2009) An integrated genetic and cytogenetic map of the cucumber genome. PLoS ONE 4:e5795
Roque A, Adsuar J (1939) New cucumber varieties resistant to the downy mildew. In: Annual report agricultural experiment station of Puerto Rico fiscal year 1937–1938, pp 45–46
Sakata Y, Kubo N, Morishita M et al (2006) QTL analysis of powdery mildew resistance in cucumber (Cucumis sativus L.). Theor Appl Genet 112:243–250
Savory EA, Granke LL, Quesada-Ocampo LM et al (2011) The cucurbit downy mildew pathogen Pseudoperonospora cubensis. Mol Plant Pathol 12:217–226
Singh V, Khan A, Saxena R et al (2016a) Next-generation sequencing for identification of candidate genes for Fusarium wilt and sterility mosaic disease in pigeonpea (Cajanus cajan). Plant Biotechnol J 14:1183–1194
Singh V, Khan A, Jaganathan D et al (2016b) QTL-seq for rapid identification of candidate genes for 100-seed weight and root/total plant dry weight ratio under rainfed conditions in chickpea. Plant Biotechnol J 14:2110–2119
Smith P (1948) Powdery mildew resistance in cucumber. Phytopathology 38:1027–1028
Szczechura W, Staniaszek M, Klosinska U, Kozik E (2015) Molecular analysis of new sources of resistance to Pseudoperonospora cubensis (Berk. et Curt.) Rostovzev in cucumber. Russ J Genet 51:974–979
Takagi H, Abe A, Yoshida K et al (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74:174–183
Van Damme M, Zeilmaker T, Elberse J et al (2009) Downy mildew resistance in Arabidopsis by mutation of HOMOSERINE KINASE. Plant Cell 21:2179–2189
Van Ooijen JW (2006) Joinmap 4.0. Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wagenigen, p 63
Van Vliet GJA, Meysing WD (1974) Inheritance of resistance to Pseudoperonospora cubensis Rost. in cucumber (Cucumis sativus L.). Euphytica 23:251–255
Van Vliet GJA, Meysing WD (1977) Relation in the inheritance of resistance to Pseudoperonospora cubensis Rost and Sphaerotheca fuliginea Poll. in cucumber (Cucumis sativus L.). Euphytica 26:793–796
VandenLangenberg KM (2015) Studies on downy mildew resistance in cucumber (Cucumis sativus L.). Dissertation, North Carolina State University
Varshney RK, Terauchi R, McCouch SR (2014) Harvesting the promising fruits of genomics: applying genome sequencing technologies to crop breeding. PLoS Biol 12:e1001883
Wan H, Yuan W, Bo K et al (2013) Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genom 14:109
Wang S, Basten CJ, Zeng ZB (2007) Windows QTL cartographer 2.5. Raleigh, NC: Department of Statistics, North Carolina State University http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Wang Y, VandenLangenberg KM, Wehner TC et al (2016) QTL mapping for downy mildew resistance in cucumber inbred line WI7120 (PI 330628). Theor Appl Genet 129:1493–1505
Wang L, Liu Z, Zhang Y et al (2018a) Identification and fine mapping of a stay-green gene (Brnye1) in pakchoi (Brassica campestris L. ssp. chinensis). Theor Appl Genet 131:673–684
Wang Y, VandenLangenberg K, Wen C et al (2018b) QTL mapping of downy and powdery mildew resistances in PI 197088 cucumber with genotyping-by-sequencing in RIL population. Theor Appl Genet 131:597–611
Wei L, Jian H, Lu K et al (2016a) Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol J 14:1368–1380
Wei Q, Fu W, Wang Y et al (2016b) Rapid identification of fruit length loci in cucumber (Cucumis sativus L.) using next-generation sequencing (NGS)-based QTL analysis. Sci Rep 6:27496
Weng Y, Johnson S, Staub JE et al (2010) An extended microsatellite genetic map of cucumber, Cucumis sativus L. HortScience 45:880–886
Wenger JW, Schwartz K, Sherlock G (2010) Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet 5:e1000942
Win K, Vegas J, Zhang C et al (2017) QTL mapping for downy mildew resistance in cucumber via bulked segregant analysis using next-generation sequencing and conventional methods. Theor Appl Genet 130:199–211
Xu X, Yu T, Xu R et al (2016) Fine mapping of a dominantly inherited powdery mildew resistance major-effect QTL, Pm1.1, in cucumber identifies a 41.1 kb region containing two tandemly arrayed cysteine-rich receptor-like protein kinase genes. Theor Appl Genet 129:507–516
Xue H, Shi T, Wang F et al (2017) Interval mapping for red/green skin color in Asian pears using a modified QTL-seq method. Hortic Res 4:17053
Yang L, Koo DH, Li Y et al (2012) Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J 71:895–906
Yang L, Li D, Li Y et al (2013) A 1,681-locus consensus genetic map of cultivated cucumber including 67 NB-LRR resistance gene homolog and ten gene loci. BMC Plant Biol 13:53
Yoshioka Y, Sakata Y, Sugiyama M, Fukino N (2014) Identification of quantitative trait loci for downy mildew resistance in cucumber (Cucumis sativus L.). Euphytica 198:265–276
Zhang H, Wang Z, Mao A et al (2008) SSR markers linked to the resistant gene of cucumber powdery mildew. Acta Agric Boreali-Sin 23:77–80 (in Chinese)
Zhang S, Liu M, Miao H et al (2011) QTL mapping of resistance genes to powdery mildew in cucumber. Sci Agric Sin 44:3584–3593
Zhang S, Liu M, Miao H et al (2013) Chromosomal mapping and QTL analysis of resistance to downy mildew in Cucumis sativus. Plant Dis 97:245–251
Zhong C, Sun S, Li P et al (2018) Next-generation sequencing to identify candidate genes and develop diagnostic markers for a novel Phytophthora resistance gene, RpsHC18, in soybean. Theor Appl Genet 131:525–538
Acknowledgements
The authors thank Martin Kagiki Njogu (Department of Horticulture, College of Horticulture, Nanjing Agricultural University, Nanjing, China) for critical reading of the manuscript. This research was supported by National Natural Science Foundation of China (Key Program, No. 31430075), Special Fund for Agro-Scientific Research in the Public Interest (No. 201403032), National Key Research and Development Program of China (2016YFD0101705-5), National Key Research and Development Program of China (2016YFD0100204-25), National Natural Science Foundation of China (No. 31672168), Independent Innovation of Agricultural Science and Technology of Jiangsu Province (CX(17)3016), Fundamental Research Funds for the Central Universities (No. KYZ201828).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical standards
The experiments were performed in accordance with all relevant Chinese laws.
Additional information
Communicated by Michael J. Havey.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, K., Wang, X., Zhu, W. et al. Complete resistance to powdery mildew and partial resistance to downy mildew in a Cucumis hystrix introgression line of cucumber were controlled by a co-localized locus. Theor Appl Genet 131, 2229–2243 (2018). https://doi.org/10.1007/s00122-018-3150-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-018-3150-2



