Abstract
Key message
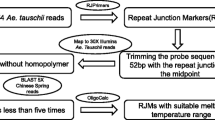
Making use of wheat chromosomal resources, we developed 11 gene-associated markers for the region of interest, which allowed reducing gene interval and spanning it by four BAC clones.
Abstract
Positional gene cloning and targeted marker development in bread wheat are hampered by high complexity and polyploidy of its nuclear genome. Aiming to clone a Russian wheat aphid resistance gene Dn2401 located on wheat chromosome arm 7DS, we have developed a strategy overcoming problems due to polyploidy and enabling efficient development of gene-associated markers from the region of interest. We employed information gathered by GenomeZipper, a synteny-based tool combining sequence data of rice, Brachypodium, sorghum and barley, and took advantage of a high-density linkage map of Aegilops tauschii. To ensure genome- and locus-specificity of markers, we made use of survey sequence assemblies of isolated wheat chromosomes 7A, 7B and 7D. Despite the low level of polymorphism of the wheat D subgenome, our approach allowed us to add in an efficient and cost-effective manner 11 new gene-associated markers in the Dn2401 region and narrow down the target interval to 0.83 cM. Screening 7DS-specific BAC library with the flanking markers revealed a contig of four BAC clones that span the Dn2401 region in wheat cultivar ‘Chinese Spring’. With the availability of sequence assemblies and GenomeZippers for each of the wheat chromosome arms, the proposed strategy can be applied for focused marker development in any region of the wheat genome.


Similar content being viewed by others
References
Azhaguvel P, Rudd JC, Ma Y, Luo MC, Weng Y (2012) Fine genetic mapping of greenbug aphid-resistance gene Gb3 in Aegilops tauschii. Theor Appl Genet 124:555–564
Berkman PJ, Skarshewski A, Lorenc M, Lai K, Duran C, Ling EYS, Stiller J, Smits L, Imelfort M, Manoli S, McKenzie M, Kubaláková M, Šimková H, Batley J, Fleury D, Doležel J, Edwards D (2011) Sequencing and assembly of low copy and genic regions of isolated Triticum aestivum chromosome arm 7DS. Plant Biotech J 9:768–775
Berkman PJ, Visendi P, Lee HC, Stiller J, Manoli S, Lorenc MT, Lai K, Batley J, Fleury D, Šimková H, Kubaláková M, Weining S, Doležel J, Edwards D (2013) Dispersion and domestication shaped the genome of bread wheat. Plant Biotech J 11:564–571
Bossolini E, Krattinger SG, Keller B (2006) Development of SSR markers specific for the Lr34 resistance region of wheat using sequence information from rice and Aegilops tauschii. Theor Appl Genet 113:1049–1062
Burd JD, Burton RL (1992) Characterization of plant-damage caused by the Russian wheat aphid (Homoptera, Aphididae). J Econ Entomol 85:2017–2022
Chao S, Zhang W, Akhunov E, Sherman J, Ma Y, Luo MC, Dubcovsky J (2009) Analysis of gene-derived SNP marker polymorphism in US wheat (Triticum aestivum L.) cultivars. Mol Breed 23:23–33
Close TJ, Wanamaker S, Roose ML, Lyon M (2007) HarvEST: an EST database and viewing software. Methods Mol Biol 406:161–178
Close TJ, Bhat PR, Lonardi S, Wu Y, Rostoks N, Ramsay L, Druka A, Stein N, Svensson JT, Wanamaker S, Bozdag S, Roose ML, Moscou MJ, Chao S, Varshney RK, Szucs P, Sato K, Hayes PM, Matthews DE, Kleinhofs A, Muehlbauer GJ, DeYoung J, Marshall DF, Madishetty K, Fenton RD, Condamine P, Graner A, Waugh R (2009) Development and implementation of high-throughput SNP genotyping in barely. BMC Genom 10:582
Collins MB, Haley SD, Peairs FB, Rudolph JB (2005) Biotype 2 Russian Wheat Aphid resistance among wheat germplasm accessions. Crop Sci 45:1877–1880
Doležel J, Vrána J, Šafář J, Bartoš J, Kubaláková M, Šimková H (2012) Chromosomes in the flow to simplify genome analysis. Funct Integr Genomics 12:397–416
Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2010) Geneious v5.3. http://www.geneious.com
FAOSTAT (2013) FAOSTAT, Food and Agriculture Organization of the United Nations. http://faostat.fao.org/
Fazel-Najafabadi M, Peng J, Peairs FB, Simkova H, Kilian A, Lapitan NLV (2014) Genetic mapping of resistance to Diuraphis noxia (Kurdjumov) Biotype 2 in wheat (Triticum aestivum L.) accession CI2401. Euphytica. doi:10.1007/s10681-014-1284-0
Girma M, Wilde GE, Harvey TL (1993) Russian wheat aphid (Homoptera: Aphididae) affects yield and quality of wheat. J Econ Entomol 86:594–601
Keller B, Feuillet C, Yahiaoui N (2005) Map-based isolation of disease resistance genes from bread wheat: cloning in a supersize geonome. Genet Res 85:93–100
Kosambi D (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Kubaláková M, Vrána J, Číhalíková J, Šimková H, Doležel J (2002) Flow karyotyping and chromosome sorting in bread wheat (Triticum aestivum L.). Theor Appl Genet 104:1362–1372
Kubaláková M, Valárik M, Bartoš J, Vrána J, Číhalíková J, Molnár-Láng M, Doležel J (2003) Analysis and sorting of rye (Secale cereale L.) chromosomes using flow cytometry. Genome 46:893–905
Lam ET, Hastie A, Lin C, Ehrlich D, Das SK, Austin MD, Deshpande P, Cao H, Nagarajan N, Xiao M, Kwok PY (2012) Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol 30:771–776
Lapitan NLV, Peng JH, Sharma V (2007) A high-density map and PCR markers for Russian wheat aphid resistance gene Dn7 on chromosome 1RS/1BL. Crop Sci 47:809–818
Liu XM, Smith CM, Gill BS, Tolmay V (2001) Microsatellite markers linked to six Russian wheat aphid resistance genes in wheat. Theor Appl Genet 102:504–510
Liu XM, Smith CM, Gill BS (2002) Identification of microsatellite markers linked to Russian wheat aphid resistance genes Dn4 and Dn6. Theor Appl Genet 104:1042–1048
Luo MC, Gu YQ, You FM, Deal KR, Ma Y, Hu Y, Huo N, Wang Y, Wang J, Chen S, Jorgensen CM, Zhang Y, McGuire PE, Pasternak S, Stein JC, Ware D, Kramer M, McCombie WR, Kianian SF, Martis MM, Mayer KF, Sehgal SK, Li W, Gill BS, Bevan MW, Šimková H, Doležel J, Weining S, Lazo GR, Anderson OD, Dvorak J (2013) A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc Natl Acad Sci USA 110:7940–7945
Lysák MA, Číhalíková J, Kubaláková M, Šimková H, Künzel G, Doležel J (1999) Flow karyotyping and sorting of mitotic chromosomes of barley (Hordeum vulgare L.). Chrom Res 7:431–444
Marais GF, Wessels WG, Horn M (1998) Association of a stem rust resistance gene (Sr45) and two Russian wheat aphid resistance genes (Dn5 and Dn7) with mapped structural loci in common wheat. S Afr J Plant Soil 15:67–71
Martin TJ, Sears RG, Seifers DL, Harvey TL, Witt MD, Schlegel AJ, McCluskey PJ, Hatchett JH (2001) Registration of Trego wheat. Crop Sci 41:929–930
Martis MM, Zhou R, Haseneyer G, Schmutzer T, Vrána J, Kubaláková M, König S, Kugler KG, Scholz U, Hackauf B, Korzun V, Schön CC, Doležel J, Bauer E, Mayer KFX, Stein N (2013) Reticulate evolution of the rye genome. Plant Cell 25:3685–3698
Mayer KFX, Taudien S, Martis M, Šimková H, Suchánková P, Gundlach H, Wicker T, Petzold A, Felder M, Steuernagel B, Scholz U, Graner A, Platzer M, Doležel J, Stein N (2009) Gene content and virtual gene order of barley chromosome 1H. Plant Physiol 151:496–505
Mayer KFX, Martis M, Hedley PE, Šimková H, Liu H, Morris JA, Steuernagel B, Taudien S, Roessner S, Gundlach H, Kubaláková M, Suchánková P, Murat F, Felder M, Nussbaumer T, Graner A, Salse J, Endo T, Sakai H, Tanaka T, Itoh T, Sato K, Platzer M, Matsumoto T, Scholz U, Doležel J, Waugh R, Stein N (2011) Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 23:1249–1263
Michalak de Jimenez MK, Bassi FM, Ghavami F, Simons K, Dizon R, Seetan RI, Alnemer LM, Denton AM, Doğramaci M, Šimková H, Doležel J, Seth K, Luo MC, Dvorak J, Gu YQ, Kianian SF (2013) A radiation hybrid map of chromosome 1D reveals synteny conservation at a wheat speciation locus. Funct Integr Genomics 13:19–32
Molnár I, Kubaláková M, Šimková H, Cseh A, Molnár-Láng M, Doležel J (2011) Chromosome isolation by flow sorting in Aegilops umbellulata and Ae. comosa and their allotetraploid hybrids Ae. biuncialis and Ae. geniculata. PLoS One 6:e27708
Morrison WP, Peairs FB (1998) Response model concept and economic impact. In: Thomas Say Publications in Entomology, Entomological Society of America, Lanham, MD
Nkongolo KK, Quick JS, Limin AE, Fowler DB (1991) Sources and inheritance of resistance to Russian wheat aphid in Triticum species amphiploids and Triticum tauschii. Can J Plant Sci 71:703–708
Peng JH, Bai Y, Haley SD, Lapitan NLV (2009) Microsatellite-based molecular diversity of bread wheat germplasm and association mapping of wheat resistance to the Russian wheat aphid. Genetica 135:95–122
Qin B, Cao A, Wang H, Chen T, You FM, Liu Y, Ji J, Liu D, Chen P, Wang XE (2011) Colinearity-based marker mining for the fine mapping of Pm6, a powdery mildew resistance gene in wheat. Theor Appl Genet 123:207–218
Quraishi UM, Abrouk M, Bolot S, Pont C, Throude M, Guilhot N, Confolent C, Bortolini F, Praud S, Murigneux A, Charmet G, Salse J (2009) Genomics in cereals: from genome-wide conserved orthologous set (COS) sequences to candidate genes for trait dissection. Funct Integr Genomics 9:473–484
Qureshi JA, Michaud JP, Martin TJ (2006) Resistance to biotype 2 Russian wheat aphid (Homoptera: Aphididae) in two wheat lines. J Econ Entomol 99:544–550
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Šafář J, Šimková H, Kubaláková M, Číhalíková J, Suchánková P, Bartoš J, Doležel J (2010) Development of chromosome-specific BAC resources for genomics of bread wheat. Cytogenet Genome Res 129:211–223
Schnurbusch T, Collins NC, Eastwood RF, Sutton T, Jefferies SP, Langridge P (2007) Fine mapping and targeted SNP survey using rice–wheat gene colinearity in the region of the Bo1 boron toxicity tolerance locus of bread wheat. Theor Appl Genet 115:451–461
Šimková H, Svensson JT, Condamine P, Hřibová E, Suchánková P, Bhat PR, Bartoš J, Šafář J, Close TJ, Doležel J (2008) Coupling amplified DNA from flow-sorted chromosomes to high-density SNP mapping in barley. BMC Genom 9:294
Šimková H, Šafář J, Kubaláková M, Suchánková P, Číhalíková J, Robert-Quatre H, Azhaguvel P, Weng Y, Peng J, Lapitan NL, Ma Y, You FM, Luo MC, Bartoš J, Doležel J (2011) BAC libraries from wheat chromosome 7D: efficient tool for positional cloning of aphid resistance genes. J Biomed Biotechnol 2011:302543
Sorrells ME, La Rota M, Bermudez-Kandianis CE, Greene RA, Kantety R, Munkvold JD, Miftahudin, Mahmoud A, Ma X, Gustafson PJ, Qi LL, Echalier B, Gill BS, Matthews DE, Lazo GR, Chao S, Anderson OD, Edwards H, Linkiewicz AM, Dubcovsky J, Akhunov ED, Dvorak J, Zhang D, Nguyen HT, Peng J, Lapitan NL, Gonzalez-Hernandez JL, Anderson JA, Hossain K, Kalavacharla V, Kianian SF, Choi DW, Close TJ, Dilbirligi M, Gill KS, Steber C, Walker-Simmons MK, McGuire PE, Qualset CO (2003) Comparative DNA sequence analysis of wheat and rice genomes. Genome Res 13:1818–1827
Terracciano I, Maccaferri M, Bassi F, Mantovani P, Sanguineti MC, Salvi S, Šimková H, Doležel J, Massi A, Ammar K, Kolmer J, Tuberosa R (2013) Development of COS-SNP and HRM markers for haplotyping and marker-assisted selection of Lr14 in durum wheat (Triticum durum Desf.). Theor Appl Genet 26:1077–1101
The International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788
Vakhrusheva OA, Nedospasov SA (2011) System of innate immunity in plants. Mol Biol 45:16–23
Valdez VA, Byrne PF, Lapitan NLV, Peairs FB, Bernardo A, Bai G, Haley SD (2012) Inheritance and genetic mapping of Russian wheat aphid resistance in iranian wheat landrace accession PI 626580. Crop Sci 52:676–682
Van Ooijen JW, Voorrips RE (2001) JoinMap 3.0, software for the calculation of genetic linkage maps. Plant J 3:739–744
Acknowledgments
We thank Prof. B.S. Gill and Prof. Adam Lukaszewski for providing seeds of the wheat ditelosomic lines. We are grateful to Hong Wang and Jeff Rudolph for RWA screening of the mapping population and Jarmila Číhalíková, Romana Šperková, Zdeňka Dubská and Jana Dostálová for the assistance with chromosome sorting and BAC library screening, and Dr. Frank Peairs for the use of the CSU Insectary and for providing aphids. We also thank Andreas Petzold and Stefan Taudien for 454 sequencing and assembling BAC clones. This work was supported by the Czech Science Foundation (Award No. P501/12/2554), Ministry of Education, Youth and Sports of the Czech Republic (National Program of Sustainability I, Grant award LO1204), and Australian Research Council (Projects LP0882095, LP0883462 and DP0985953). Partial support for RWA screening was provided by USDA Cooperative Agreement no. 2010-34205-21350.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H.-Q. Ling.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Staňková, H., Valárik, M., Lapitan, N.L.V. et al. Chromosomal genomics facilitates fine mapping of a Russian wheat aphid resistance gene. Theor Appl Genet 128, 1373–1383 (2015). https://doi.org/10.1007/s00122-015-2512-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2512-2