Abstract
In contrast to most autotrophic plants, which produce carbohydrates from carbon dioxide using photosynthesis, parasitic weed plants rely on host plants to form vascular connections through which they withdraw the required nutritive resources and water. Many important crop plants are infested by these heterotrophic plants leading to tremendous yield losses and rendering agricultural lands uncultivable. The parasitic weeds are physically attached to the host plants and therefore their control is challenging due to the lack of selective methods for killing the weeds without damaging the host crop. Fortunately, many host plants have pre-haustorium resistance, host initiation responses and post-attachment tolerance to these parasitic weeds. However, parasitic weeds have high fecundity, dispersal efficiency, and persistent seed storage in the soil all of which enable them to adapt to new hosts and break down the crop resistance. Recent discoveries in genome editing and gene silencing-based technologies open new opportunities to enhance crop resistance to parasitic weeds. Some genome editing-based studies targeting the seed germination of parasitic weeds created almost complete resistance in crop species. In this chapter, we give an overview of the host-parasitic interaction and host defence responses that can be targeted by genome editing or gene silencing technologies.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
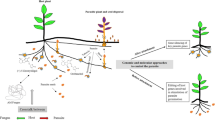
Plants are autotrophic organisms using light as energy to convert inorganic carbon into carbohydrates by photosynthesis. However, some plants have evolved specialized organs (haustorium) to attach and form vascular connections with autotrophic plants to absorb their water and nutrients. This heterotrophic lifestyle is used by parasitic plants/weeds and has a profound negative impact on many agriculturally important crops, forests and whole dynamics of ecological systems [1]. Parasitic plants could be grouped as facultative or obligatory according to their dependency on the host. Facultative parasitic plants (hemiparasitic) have their own chlorophyll and can complete their life cycle independently of a host. However, if there is an available host plant to obtain nutrients and water with less investment in the assimilation system, they become parasitic. Obligate parasitic plants (holoparasites) lack chlorophyll and they depend completely on their hosts for seed germination and survival. Parasitic plants can also be separated as root feeders or shoot feeders based on the invaded host tissue. Depending on their vascular connections with their host, they could be xylem feeders, phloem feeders, or both [2].
Parasitic plants in lower diversified agricultural systems can cause tremendous yield losses rendering agricultural lands uncultivable [3]. Traditional control methods such as hand weeding and herbicide treatment are too expensive and labour-intensive to regulate parasitic plant infestations in crops. These methods also are ineffective due to the tight physiological connection between the host and the parasitic weed and the re-emergence of parasitic plants after damaging of the host. Parasitic plants generally produce plentiful small seeds contaminating the soil or the crop seeds before parasitism is established. The seeds of parasitic plants remain viable in the dormant state for many years and germinate after receiving the host signals [4].
Reducing the impact and spread of parasitic weeds on crops and agricultural production requires an understanding of the molecular machinery behind the interactions between the parasite and the host plants. Pre and post-attachment as well as haustorium initiation resistance mechanisms in specific cultivars, mutants, or species have been identified and many host metabolites required for the germination of parasitic weed seeds have been identified. The availability of whole-genome sequences and transcriptomes of several parasitic plants facilitated the investigation of genes responsible for host–parasite interactions, and the identification of the genes involved in resistance or susceptibility responses of crops [5,6,7]. All this knowledge can be used to enhance resistance in crop species to these weeds by deploying molecular breeding and advanced genome editing strategies. In this chapter, we provide a comprehensive overview of new genome editing or gene silencing-based approaches applied to crops to enhance parasitic weed resistance and their prospective applications on the molecular mechanisms involved in host-parasitic weed interaction.
2 Genome Editing-Based Strategies Used to Enhance Parasitic Weeds Resistance in Crops
Many agriculturally important plants are attacked by specific parasitic plants, which induces a host defence response to inhibit the attachment of parasitic weeds or reduce the infestation. Based on whether the resistance mechanism functions before or after parasitic plants attach to their hosts, resistance responses can be classified as pre-haustorium resistance, haustorium initiation resistance or post-attachment resistance [8]. In addition to the classical transgenic approach, newly discovered biotechnological strategies (RNAi, VİGS and CRISPR) have been implemented to develop a high level of crop resistance to parasitic weeds in recent years [9,10,11]. In the review, we grouped these studies according to their target resistance mechanisms.
2.1 Genome Editing for Pre-HAUSTORIUM Resistance in Crops
The discovery of some terpenoid lactones in crops such as strigolactones (SLs) and sesquiterpene lactones (STLs), [11, 12] is a milestone in understanding the interaction between parasitic weeds and their hosts. Secondary metabolites synthesized by host roots in trace amounts have several important physiological processes in host plants from shoot branching to arbuscular mycorrhizal symbiosis. Terpenoid lactones were then realized to be also the germination stimulants for several obligate root parasitic plants [13, 14]. The seeds of these parasitic plants do not germinate unless they receive terpenoid lactones as a chemical signal from their host roots. Therefore, the parasite-host interaction has evolved in a sophisticated way to detect the presence of STLs or SLs by parasitic weeds and coordinate their germination and development with the host’s lifecycle [15, 16]. Receiving the signal molecule from the host for seed germination and growth towards the host organs are critical steps in the parasitic plant life cycle. Resistant host plants take a preventive pre-attachment strategy by making themselves invisible to parasitic plants by decreasing or completely stopping the production of germination stimulant molecules [17]. Therefore, reducing the number of stimulants exuded by host plants is considered to be a key factor for the host resistance achieved by inhibition of parasitic weed seed germination. Clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9 (CRISPR/Cas9)-mediated mutagenesis, virus-induced gene silencing (VIGS) and RNA interference (RNAi) mediated gene silencing strategies have been used to disrupt strigolactones (SLs) biosynthesis in host plants [18,19,20,21,22,23]. In this way, the germination of seeds of parasitic plants was suppressed and almost complete resistance to parasitic weeds was achieved in genome-edited host plants (Table 24.1).
SLs were first isolated in cotton root exudates as a germination stimulant of Striga lutea [24]. Subsequent research revealed that these compounds also function as endogenous hormones to inhibit shoot branching or tillering. SL biosynthesis begins with the convertion of all-trans-β-carotene to 9-cis-β-carotene with an enzyme called β-carotene isomerase (DWARF27 or D27). Then, carotenoid cleavage dioxygenase 7 (CCD7) cleaves 9-cis-β-carotene into the volatile β-ionone and 9-cis-β-apo-10′-carotenal. This former intermediate is catalyzed by CCD8 to yield carlactone which is the precursor for all SLs (Fig. 24.1). In Arabidopsis thaliana, carlactone is converted into carlactonoic acid by the cytochrome P450 monooxygenase (MORE AXILLARY GROWTH 1-MAX1) (Fig. 24.1a). In rice, MAX1 homologs convert carlactone into 4-deoxyorobanchol and orobanchol [25].
Strigolactones (SLs) released from the host roots are the main stimulants for seed germination of parasitic weeds. Therefore the genes encoding the enzymes functional in SL biosynthesis (a) were the targets for CRISPR-mediated gene knockout studies. CRISPR-mediated disruption of the CC7, CCD8 and MAX1 genes in rice (b) and tomato (c, d) to reduce SL content in the root exudates. All the SL-deficient mutant plants exhibited reduced or poor germination in the seeds of parasitic plants such as S. hermonthica, O. crenata and P. aegyptiaca
RNA interference (RNAi) uses an antisense siRNA strand to associate with the RNA-induced silencing complex (RISC) to target homologous RNA molecules for degradation and gene silencing in plants [26]. RNAi was previously used to silence several key genes encoding critical enzymes functional in SL biosynthesis. Gene silencing of CCD7 and CCD8 transcripts in tomatoes using antisense siRNA resulted in decreased levels of SL in the host, leading to reduced germination of the root parasitic weed [18, 19, 27].
Kohlen et al. (2012) showed that silencing of the host CCD8 gene in tomato lines by hpRNA technique reduces infestation of P. ramosa by 90% in the transgenic plants [19]. In another study, Aly et al. (2014) used a tobacco rattle virus–VIGS system for the transient knockdown of CCD7 or CCD8 in P. aegyptiaca. The result of the study demonstrated significant inhibition of parasite-tubercle development and the infestation of Nicotiana benthamiana plants [20]. A similar approach was used for the control of root parasitic weeds based on the simultaneous trans-specific gene silencing of parasite genes [29]. In this study, multiple DNA fragments (ACS, M6PR, and Prx1) of P. aegyptiaca genes were targeted by RNAi. The results of the experiment showed the movement of mobile exogenous siRNA from the host to the parasite, which lead to the decreased expression of parasitic genes essential for the parasite tubercles growing on the host plants.
CRISPR/Cas9 is the newest genome editing approach used to silence or modify the genes of plant species to enhance resistance to parasitic plants. This efficient and simple genome editing tool requires a small-guided RNA (sgRNA) complementary to a target gene sequence and Cas9 enzyme that recognize sgRNA for precise cutting of DNA and leading to dsDNA breaks [30]. During DNA repair by non-homologous end joining, insertion or deletion may occur at the break sites, silencing the protein’s function [31]. CRISPR/Cas9 has been recently applied to knock out the CCD7 gene in rice (Oryza sativa) to reduce SL content in the roots [21]. CCD7 mutants exhibited increased tillering, combined with reduced height and extremely poor levels of SL production compared to the wild-type control. Striga seed germination was almost completely inhibited by the root exudates of some CCD7 mutants compared to that of control and the standard SL analogue GR24 (Fig. 24.1b). In another study, CRISPR/Cas9-mediated mutagenesis of the CCD8 gene was used to enhance host resistance to the parasitic weed P. aegyptiaca [22]. In this study, Cas9/single guide RNA constructs were targeted to the second exon of CCD8 in tomato plants. Several mutant tomato lines with heritable insertions or deletions in CCD8 gene were recorded to be SL-deficient. Compared to control tomato plants, the CCD8 mutant lines had morphological changes such as dwarfing, excessive shoot branching and adventitious root formation. In addition, some SL-deficient CCD8 mutants exhibited an almost complete reduction in seed germination of P. aegyptiaca and its infestation compared to non-mutated tomato plants (Fig. 24.1c) [22]. Wakabayashi et al. (2019) knocked out the cytochrome P450 (MAX1) gene, SlCYP722C, coding for an orobanchol synthase enzyme in tomato, by using a CRISPR system (Fig. 24.1d) [23]. Indels in the gene that resulted in biallelic frameshift mutations were identified in the T1 transgenic plants and T2 progeny lines. Orobanchol production was completely inhibited in the root exudates of MAX1 mutant tomato plants. Unlike the CCD8 mutant tomato lines created by Bari et al. [22], MAX1 mutants did not show prominent phenotypes such as increased shoot branching and reduced stem length. Production of the fruits and seeds was normal in the T1 MAX1 mutant tomato lines normally, and no serious yield loss occurred in mutant T2 progeny. Most importantly, root exudates of MAX1 mutant tomato plants reduced the induction of germination of seeds of root parasitic weeds, Striga hermonthica, Orobanche crenata, and Phelipanche aegyptiaca, compared to WT without changing the plant architecture.
Secretion of toxic compounds inhibiting the seed germination of parasitic weeds is another strategy for host resistance against parasitic weeds. Many phytotoxins or natural amino acids were found to interfere with the early growth stages of the parasitic weeds. These metabolites have negative effects on seed germination or germ tube elongation [32]. Serghini et al. (2001) found that the resistant sunflower genotypes release defensive secondary metabolites called 7-hydroxylated coumarins from their root to create a toxic environment for O. cernua [33]. In another study, transgenic tobacco overexpressing an antibacterial peptide sarcotoxin IA enhanced resistance to Phelipanche spp. by its toxic effects on this parasitic weed [34].
2.1.1 Genome Editing for HAUSTORIUM Initiation Resistance in Crops
Once a germination signal is released from the host and detected by the parasitic plants, a haustorium contact is established between the host and the parasite. Therefore, instead of reducing the germination of parasitic seeds, inhibition of haustorium formation via several Haustorium Induction Factors (HIFs) could be also another strategy for crop resistance. HIFs are released from the parasitic plants to enable haustoria penetration into host organs following haustorium attachment [35]. A quinone molecule, 2,6-dimethoxy-1,4-benzoquinone (DMBQ), released from sorghum root extract was the first defined HIF molecule in the parasitic plants. DMBQ was recorded to induce both obligate and facultative parasitic haustoria development within hours [36]. Interestingly, two genes (TvQR1 and TvQR2) encoding a type of quinone oxidoreductases in Triphysaria versicolor (facultative parasitic plants) were identified to be responsible for the induction of DMBQ. TvQR1 was estimated to generate the first step in the signal-transduction pathway for haustorium development while TvQR2 was thought to be responsible for the removal of the signal with a detoxification system [37]. RNA interference (RNAi) technology was used to silence TvQR1and TvQR2 transcripts in Triphysaria roots for the evaluation of their functional role in haustoria formation. In the study, RNAi vectors designed to target TvQR1 and TvQR2 were transformed into Triphysaria roots via Agrobacterium rhizogenes. The competence of transgenic Triphysaria roots was accomplished by Arabidopsis root contact test. The results of haustoria formation in response to host contact indicated a significant decrease in haustorium development in roots silenced for QR1 but not in roots silenced for QR2. This experiment implicates QR1 as the first identified gene necessary for the redox bioactivation of haustorial-inducing factors [38].
2.1.2 Genome Editing to Enhance Post-attachment Resistance in Crops
Even after the seeds of parasitic weeds germinate and attach to the host roots, hormone-mediated defence response in host plants can be triggered to cope with this parasite attack. Defence-related plant hormones, especially jasmonic acid (JA) and salicylic acid (SA), are known to contribute to crop resistance to parasitic weeds by direct inhibition of their contact with the host or enhancing the host plant vascular body. For instance, treatment of SA on red clover roots reduced the houstaria formation of O. minor by lignification in the host endodermis cell layers [39]. Induction of SA and pathogenesis-related gene transcripts were also reported to enhance the resistance response of sunflowers to O. cumana [40]. JA is known to be involved in cell wall damage–induced lignin biosynthesis and, therefore, it directly contributed to the host resistance by a hypersensitive-like response in plants [35]. Several studies have concentrated on the loss of function analysis of these hormones in crop species. For instance, Brading et al. (2000) created a transgenic tomato expressing salicylate hydroxylase. This enzyme converted SA immediately to inactive catechol and created SA-deficient tomato [41]. In another study, the radiation-based mutation was created on the tomato CORONATINE-INSENSITIVE1 gene which reduced the expression of JA-responsive genes [42]. Runyon et al. (2010) used both mutant tomato genotypes to test their resistance to parasitic weeds. The results indicated that parasitic plants grown on the SA and JA mutant tomatoes were more aggressive and had more biomass than those grown on their wild-type counterparts [43].
In another study, RNAi was used to knock down (kd) the expression JA-inducible WRKY transcription factor in rice [44]. Remarkably, WRKY45-kd rice genotypes exhibited severe susceptibility to S. hermonthica. The size and number of the S. hermonthica seedlings that attached and developed in mutant rice genotypes were almost threefold higher compared with wild-type rice. Therefore, a reduction in endogenous JA levels resulted in enhanced susceptibility to S. hermonthica. External application of JA was found to completely recover the resistance ability of mutant rice to this parasitic plant [5, 45, 46].
Hypersensitive response (HR) is a common mechanism which leads to localized cell death and necrosis at the infectious site to defend against pathogens and prevent the spread of infection in the plant body [47]. Some studies indicated that hosts have evolved the ability to detect parasitic plant–specific signals to initiate signal transduction cascades that lead to an HR and prevent the haustorium penetration process of parasitic plants [48]. For instance, a cowpea cultivar resistant to S. gesnerioides was found to trigger a downstream signalling cascade to activate the avirulence (Avr) proteins, which is a positive regulator of the HR [49]. A similar case was also reported for the interaction between sunflowers and O. cumana. Sunflower recognizes an avirulence protein (AVROR7) from O. cumana via the kinase domain of the HAOR7 protein, which then activates signalling cascades for the induction of HR [50].
3 Prospective Applications of Genome Editing-Based Systems for the Control of Parasitic Plants in Crops
Genome editing-based strategies used to silence host or parasite genes may serve as an important strategy to obtain more effective and durable crop resistance to parasitic weeds. Unlike other types of natural resistance, genome editing-based strategies could be easily applied to susceptible crop cultivars. Moreover, parasite species share homology in the target gene sequence and, therefore, an established strategy could be effective against other parasitic weed species. For instance, Aly et al. (2009) revealed that M6PR gene has high sequence similarity between P. aegyptiaca, P. ramosa and O. crenata species, suggesting that a single RNAi or CRISPR-based protocol can be used to manipulate sensitivity to several species at the same time. In addition, multiple candidate parasitic genes can be cloned in the same construct and pyramided in susceptible hosts for gene editing, thus significantly reducing the risk of the development of new virulent parasitic weeds [28]. A limited number of studies described in-vitro transformation and regeneration systems for P. aegyptiaca [51]. and P. ramosa [52]. However, the establishment of genome editing-based protocols needs more effort to acquire high-quality genomic data, reverse-genetics information and reliable parasite transformation systems to target key processes in the host–parasite interaction. Nevertheless, current molecular knowledge could still be a target for CRISPR-based genome editing studies. For instance, RNAi or CRISPR-based silencing of parasite genes functional in host cell-wall degradation and penetration (pectin methylesterase, polygalacturonase, rhamnogalacturonase or peroxidases) may reduce host penetrability during haustorium formation and initial parasitic stages. Another promising strategy could be the reduction of the parasite’s seed productivity by silencing the genes involved in flower and seed formation pathways. A general conclusion emerging from research in the last 20 years is that the intimate physical and physiological connection of parasites with their hosts can be used as a key target point, where its greatest potential lies in developing parasite resistance utilizing molecule or macromolecule exchange. Since host-released stimulants such as hormones, seconder metabolites and signals are the most critical key factor in germination and infestation of parasitic weeds, genes involved in these stimulant biosynthesis communications, signalling, and perception should be further studied and identified for the best targets for genome editing. A more thorough understanding of molecular interaction between host and parasite will enable manipulation of their in-vivo interactions and activity to control root parasitic weed germination without damaging the crop plant.
References
Hu, L., Wang, J., Yang, C., Islam, F., Bouwmeester, H.J., et al.: The effect of virulence and resistance mechanisms on the interactions between parasitic plants and their hosts. Int. J. Mol. Sci. 21, 9013 (2020)
Fernández-Aparicio, M., Delavault, P., Timko, M.P.: Management of infection by parasitic weeds: a review. Plants (Basel). 9(9), 1184 (2020)
Delavault, P.: Are root parasitic plants like any other plant pathogens? New Phytol. 226, 641–643 (2020)
Mutuku, J.M., Cui, S., Yoshida, S., Shirasu, K.: Orobanchaceae parasite–host interactions. New Phytol. 230, 46–59 (2021)
Yıldırım, K., Kavas, M., Kaya, R., Seçgin, Z., Can, C., Sevgen, I., Saraç, G., Tahan, V.: Genome-based identification of beet curly top Iran virus infecting sugar beet in Turkey and investigation of its pathogenicity by agroinfection. J. Virol. Methods. 300, 114380 (2021)
Fishman, M.R., Shirasu, K.: How to resist parasitic plants: pre- and post-attachment strategies. Curr. Opin. Plant Biol. 62, 102004 (2021)
Panstruga, R., Moscou, M.J.: What is the molecular basis of nonhost resistance? Mol. Plant-Microbe Interact. 33, 1253–1264 (2020)
Aksoy, E., Yildirim, K., Kavas, M., Kayihan, C., Yerlikaya, B.A., Calik, I., Sevgen, I., Demirel, U.: General guidelines for CRISPR/Cas-based genome editing in plants. Mol. Biol. Rep. 49, 12151–12164 (2022)
Yıldırım, K., Kavas, M., Küçük, İ.S., Seçgin, Z., Saraç, Ç.G.: Development of highly efficient resistance to Beet Curly Top Iran Virus (Becurtovirus) in Sugar Beet (B. vulgaris) via CRISPR/Cas9 system. Int. J. Mol. Sci. 24(7), 6515 (2023)
Miladinovic, D., Antunes, D., Yildirim, K., Bakhsh, A., Cvejić, S., Kondić-Špika, A., Jeromela, A.M., Opsahl-Sorteberg, H.G., Zambounis, A., Hilioti, Z.: Targeted plant improvement through genome editing: from laboratory to field. Plant Cell Rep. 40, 935–951 (2021)
Xie, X., Yoneyama, K., Yoneyama, K.: The strigolactone story. Annu. Rev. Phytopathol. 48, 93–117 (2010)
Chadwick, M., Trewin, H., Gawthrop, F., Wagstaff, C.: Sesquiterpenoids lactones: benefits to plants and people. Int. J. Mol. Sci. 14(6), 12780–12805 (2013)
Cheng, X., Floková, K., Bouwmeester, H., Ruyter-Spira, C.: The role of endogenous strigolactones and their interaction with ABA during the infection process of the parasitic weed Phelipanche ramosa in tomato plants. Front. Plant Sci. 8, 392 (2017)
Raupp, F.M., Spring, O.: New sesquiterpene lactones from sunflower root exudate as germination stimulants for Orobanche cumana. J. Agri. Food Chem. 61, 10481–10487 (2013)
López-Ráez, J.A., Matusova, R., Cardoso, C., Jamil, M., Charnikhova, T., et al.: Strigolactones: ecological significance and use as a target for parasitic plant control. Pest Manag. Sci. 65, 471–477 (2009)
Spring, O.: Sesquiterpene lactones in sunflower oil. LWT. 142, 111047 (2021)
Ashapkin, V.V., Kutueva, L.I., Aleksandrushkina, N.I., Vanyushin, B.F., Teofanova, D.R., Zagorchev, L.I.: Genomic and epigenomic mechanisms of the interaction between parasitic and host plants. Int. J. Mol. Sci. 24, 2647 (2023)
Vogel, J.T., Walter, M.H., Giavalisco, P., Lytovchenko, A., Kohlen, W., Charnikhova, T., Simkin, A.J., Goulet, C., Strack, D., Bouwmeester, H.J., et al.: SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J. 61, 300–311 (2010)
Kohlen, W., Charnikhova, T., Lammers, M., Pollina, T., Tóth, P., Haider, I., Pozo, M.J., Maagd, R.A., Ruyter-Spira, C., Bouwmeester, H.J., et al.: The tomato CAROTENOID CLEAVAGE DIOXYGENASE 8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol. 196, 535–547 (2012)
Aly, R., Dubey, N.K., Yahyaa, M., Abu-Nassar, J., Ibdah, M.: Gene silencing of CCD7 and CCD8 in Phelipanche aegyptiaca by tobacco rattle virus system retarded the parasite development on the host. Plant Signal. Behav. 9(8), e29376 (2014)
Butt, H., Jamil, M., Wang, J.Y., Al-Babili, S., Mahfouz, M.: Engineering plant architecture via CRISPR/Cas9-mediated alteration of strigolactone biosynthesis. BMC Plant Biol. 18, 174 (2018)
Bari, V.K., Nassar, J.A., Kheredin, S.M., et al.: CRISPR/Cas9-mediated mutagenesis of CAROTENOID CLEAVAGE DIOXYGENASE 8 in tomato provides resistance against the parasitic weed Phelipanche aegyptiaca. Sci. Rep. 9, e11438 (2019)
Wakabayashi, T., Hamana, M., Mori, A., et al.: Direct conversion of carlactonoic acid to orobanchol by cytochrome P450 CYP722C in strigolactone biosynthesis. Sci. Adv. 5, eaax9067 (2019)
Lee, H.A., Lee, H.Y., Seo, E., Lee, J., Kim, S.B., et al.: Current understandings of plant nonhost resistance. Mol. Plant-Microbe Interact. 30, 5–15 (2017)
Mashiguchi, K., Seto, Y., Yamaguchi, S.: Strigolactone biosynthesis, transport and perception. Plant J. 105(2), 335–350 (2021)
de Framond, A., Rich, P.J., McMillan, J., Ejeta, G.: Effects on Striga parasitism of transgenic maize armed with RNAi constructs targeting essential S. asiatica genes. In: Gebisa, E., Jonathan, G. (eds.) Integrating New Technologies for Striga Control, pp. 185–196. World Scientific, Singapore (2007)
Aly, R., Matzrafi, M., Bari, V.K.: Using biotechnological approaches to develop crop resistance to root parasitic weeds. Planta. 253(5), 97 (2021)
Aly, R., Cholakh, H., Joel, D.M., Leibman, D., Steinitz, B., Zelcer, A., Naglis, A., Yarden, O., Gal-On, A.: Gene silencing of mannose 6-phosphate reductase in the parasitic weed Orobanche aegyptiaca through the production of homologous dsRNA sequences in the host plant. Plant Biotechnol. J. 7, 487–498 (2009)
Dubey, N.K., Eizenberg, H., Leibman, D., Wolf, D., Edelstein, M., Abu-Nassar, J., Marzouk, S., Gal-On, A., Aly, R.: Enhanced host-parasite resistance based on down-regulation of Phelipanche aegyptiaca target genes is likely by mobile small RNA. Front. Plant Sci. 8, 1574 (2017)
Seçgin, Z., Kavas, M., Yildirim, K.: Optimization of agrobacterium-mediated transformation and regeneration for CRISPR/Cas9 genome editing of commercial tomato cultivars. Turk. J. Agric. For. 45, 704–716 (2021)
Secgin, Z., Uluisik, S., Yıldırım, K., Abdulla, M.F., Mostafa, K., Kavas, M.: Genome-wide identification of the Aconitase gene family in tomato (Solanum lycopersicum) and CRISPR-based functional characterization of SlACO2 on male-sterility. Int. J. Mol. Sci. 23(22), 13963 (2022)
Vurro, M., Boari, A., Evidente, A., Andolfi, A., Zermane, N.: Natural metabolites for parasitic weed management. Pest Manag. Sci. 65(5), 566–571 (2009)
Serghini, K., de Luque, A.P., Castejón-Muñoz, M., García-Torres, L., Jorrín, J.V.: Sunflower (Helianthus annuus L.) response to broomrape (Orobanche cernua Loefl.) parasitism: induced synthesis and excretion of 7-hydroxylated simple coumarins. J. Exp. Bot. 52, 2227–2234 (2001)
Hamamouch, N., Westwood, J.H., Banner, I., Cramer, C.L., Gepstein, S., Aly, R.: A peptide from insects protects transgenic tobacco from a parasitic weed. Transgenic Res. 14(3), 227–236 (2005)
Albanova, I.A., Zagorchev, L.I., Teofanova, D.R., Odjakova, M.K., Kutueva, L.I., Ashapkin, V.V.: Host resistance to parasitic plants—current knowledge and future perspectives. Plan. Theory. 12, 1447 (2023)
Goyet, V., Wada, S., Cui, S., Wakatake, T., Shirasu, K., et al.: Haustorium inducing factors for parasitic Orobanchaceae. Front. Plant Sci. 10, 1056 (2019)
Matvienko, M., Wojtowicz, A., Wrobel, R., Jamison, D., Goldwasser, Y., Yoder, J.I.: Quinone oxidoreductase message levels are differentially regulated in parasitic and non-parasitic plants exposed to allelopathic quinones. Plant J. 25(4), 375–387 (2001)
Bandaranayake, P.C., Filappova, T., Tomilov, A., Tomilova, N.B., Jamison-McClung, D., Ngo, Q., Inoue, K., Yoder, J.I.: A single-electron reducing quinone oxidoreductase is necessary to induce haustorium development in the root parasitic plant Triphysaria. Plant Cell. 22(4), 1404–1419 (2010)
Kusumoto, D., Goldwasser, Y., Xie, X., Yoneyama, K., Takeuchi, Y., Yoneyama, K.: Resistance of red clover (Trifolium pratense) to the root parasitic plant Orobanche minor is activated by salicylate but not by jasmonate. Ann. Bot. 100, 537–544 (2007)
Letousey, P., De Zelicourt, A., Vieira Dos Santos, C., Thoiron, S., Monteau, F., Simier, P., Thalouarn, P., Delavault, P.: Molecular analysis of resistance mechanisms to Orobanche cumana in sunflower. Plant Pathol. 56, 536–546 (2007)
Brading, P.A., Hammond-Kosack, K.E., Parr, A., Jones, J.D.G.: Salicylic acid is not required for Cf-2- and Cf-9-dependent resistance of tomato to Cladosporium fulvum. Plant J. 23, 305–318 (2000)
Li, L., Zhao, Y.F., McCaig, B.C., Wingerd, B.A., Wang, J.H., Whalon, M.E., Pichersky, E., Howe, G.A.: The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell. 16, 126–143 (2004)
Runyon, J.B., Mescher, M.C., Felton, G.W., De Moraes, C.M.: Parasitism by Cuscuta pentagona sequen- tially induces JA and SA defence pathways in tomato. Plant Cell Environ. 33, 290–303 (2010)
Shimono, M., Sugano, S., Nakayama, A., et al.: Rice WRKY45 plays a crucial role in Benzothiadiazole-inducible blast resistance. Plant Cell. 19(6), 2064–2076 (2007)
Mutuku, J.M., Cui, S., Hori, C., Takeda, Y., Tobimatsu, Y., et al.: The structural integrity of lignin is crucial for resistance against Striga hermonthica parasitism in rice. Plant Physiol. 179, 1796–1809 (2019)
Mutuku, J.M., Yoshida, S., Shimizu, T., Ichihashi, Y., Wakatake, T., et al.: The WRKY45-dependent signaling pathway is required for resistance against Striga hermonthica parasitism. Plant Physiol. 168, 1152–1163 (2015)
Yildirim, K., Boylu, B., Atici, E., TKahraman Akkaya, M.S.: In Turkish wheat cultivars the resistance allele of LR34 is ineffective against leaf rust. J. Plant Dis. Prot. 119, 135–141 (2012)
Saucet, S.B., Shirasu, K.: Molecular parasitic plant–host interactions. PLoS Pathog. 12, e1005978 (2016)
Su, C., Liu, H., Wafula, E.K., Honaas, L., de Pamphilis, C.W., Timko, M.P.: SHR4z, a novel decoy effector from the haustorium of the parasitic weed Striga gesnerioides, suppresses host plant immunity. New Phytol. 226(3), 891–908 (2020)
Duriez, P., Vautrin, S., Auriac, M.C., Bazerque, J., Boniface, M.C., et al.: A receptor-like kinase enhances sunflower resistance to Orobanche cumana. Nat. Plant. 5, 1211–1215 (2019)
Fernandez-Aparicio, M., Rubiales, D., Bandaranayake, P.C., Yoder, J.I., Westwood, J.H.: Transformation and regeneration of the holoparasitic plant Phelipanche aegyptiaca. Plant Methods. 7, 36 (2011)
Libiakova, D., Ruyter-Spira, C., Bouwmeester, H.J., Matusova, R.: Agrobacterium rhizogenes transformed calli of the holoparasitic plant Phelipanche ramosa maintain parasitic competence. Plant Cell Tissue Organ Cult. 135, 321–329 (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2024 The Author(s)
About this chapter
Cite this chapter
Yıldırım, K., Kavas, M., Akın, M., Küçük, İ.S. (2024). Genome Editing-Based Strategies Used to Enhance Crop Resistance to Parasitic Weeds. In: Ricroch, A., Eriksson, D., Miladinović, D., Sweet, J., Van Laere, K., Woźniak-Gientka, E. (eds) A Roadmap for Plant Genome Editing . Springer, Cham. https://doi.org/10.1007/978-3-031-46150-7_24
Download citation
DOI: https://doi.org/10.1007/978-3-031-46150-7_24
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-46149-1
Online ISBN: 978-3-031-46150-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)