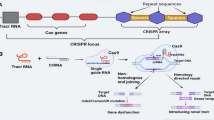
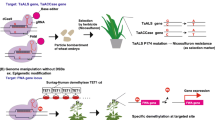
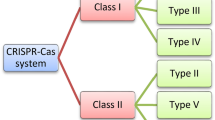
Abstract
CRISPR (clustered regularly interspaced short palindromic repeats)/Cas (CRISPR-associated) technology is a versatile genome editing tool that has been used to improve agriculturally important plant traits. Due to its precision, CRISPR/Cas9 is more effective than either conventional plant breeding methods or standard genetic engineering approaches for the rapid development of new varieties resilient to climate change. In addition to knowledge in tissue culture-based plant transformation, effective gene-specific single guide RNA (sgRNA) design, prediction of its off-target effect and utilization of vectors, promoters, Cas proteins and terminators is required for CRISPR/Cas9. Various bioinformatics tools are available for the best sgRNA design and screening of the off-targets. Various tools are used in the delivery of CRISPR/Cas components into cells and the genome. Moreover, some recent studies proved the simultaneous silencing of different paralogs in the same family or several genes working in the same pathway by using multiple-target sgRNA designs. This review summarizes the type of promoters, Cas proteins, recognition sequences, and terminators available for the development of knock-out and overexpression plant lines. It also provides a general guideline for the development of genome-edited plants from the design of sgRNAs to the selection of non-transgenic genome-edited T2 generation.

Similar content being viewed by others
Data availability
Not applicable.
References
Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346:6213. https://doi.org/10.1126/science.1258096
Makarova KS, Wolf YI, Alkhnbashi OS et al (2015) An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol 13:722–736. https://doi.org/10.1038/nrmicro3569
Zhang Y, Malzahn AA, Sretenovic S, Qi Y (2019) The emerging and uncultivated potential of CRISPR technology in plant science. Nat Plants 5:778–794. https://doi.org/10.1038/s41477-019-0461-5
Jiang F, Doudna JA (2015) The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol 30:100–111. https://doi.org/10.1016/j.sbi.2015.02.002
Westra ER, van Erp PBG, Künne T (2012) CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell 46:595–605. https://doi.org/10.1016/j.molcel.2012.03.018
Hale CR, Zhao P, Olson S (2009) RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956. https://doi.org/10.1016/j.cell.2009.07.040
Ishino Y, Krupovic M, Forterre P (2018) History of CRISPR-Cas from encounter with a mysterious repeated sequence to genome editing technology. J Bacteriol 200:e00580-e617. https://doi.org/10.1128/JB.00580-17
Zafar SA, Zaidi SSA, Gaba Y (2020) Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J Exp Bot 71:470–479. https://doi.org/10.1093/jxb/erz476
Hsu PD, Scott DA, Weinstein JA (2013) DNA targeting specificity of RNAguided Cas9 nucleases. Nat Biotechnol 31:827–832. https://doi.org/10.1038/nbt.2647
Stella S, Alcon P, Montoya G (2017) Class 2 CRISPR–Cas RNA-guided endonucleases: Swiss Army knives of genome editing. Nat Struct Mol Biol 24:882–892. https://doi.org/10.1038/nsmb.3486
Klompe SE, Vo PL, Halpin-Healy TS, Sternberg SH (2019) Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 571:219–225. https://doi.org/10.1038/s41586-019-1323-z
Niewoehner O, Garcia-Doval C, Rostøl JT (2017) Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature 548:543–548. https://doi.org/10.1038/nature23467
Makarova KS, Wolf YL, Iranzo J (2020) Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18:67–83. https://doi.org/10.1038/s41579-019-0299-x
Wada N, Osakabe K, Osakabe Y (2022) Expanding the plant genome editing toolbox with recently developed CRISPR-Cas systems. Plant Physiol 188:1825–1837. https://doi.org/10.1093/plphys/kiac027
Anzalone AV, Koblan LW, Liu DR (2020) Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38:824–844. https://doi.org/10.1038/s41587-020-0561-9
Kim H, Kim ST, Ryu J (2017) CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun 8:1–7. https://doi.org/10.1038/ncomms14406
Elmore JR, Sheppard NF, Ramia N (2016) Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR–Cas system. Genes Dev 30:447–459. https://doi.org/10.1101/gad.272153.115
Malzahn AA, Tang X, Lee K (2019) Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol 17:1–14. https://doi.org/10.1186/s12915-019-0629-5
Zhan X, Lu Y, Zhu JK, Botella JR (2020) Genome editing for plant research and crop improvement. J Integr Plant Biol 63:3–33. https://doi.org/10.1111/jipb.13063
Abudayyeh OO, Gootenberg JS, Essletzbichler P (2017) RNA targeting with CRISPR-Cas13. Nature 550:280–284. https://doi.org/10.1038/nature24049
Ali Z, Mahas A, Mahfouz M (2018) CRISPR/Cas13 as a tool for RNA interference. Trends Plant Sci 23:374–378. https://doi.org/10.1016/j.tplants.2018.03.003
Pandita D, Palakolanu PCOR, SR (2021) CRISPR/Cas13: a novel and emerging tool for RNA editing in plants. Tang G, Teotia S, Tang X, Singh D (eds) RNA-based technologies for functional genomics in plants. Springer, Cham, pp 301–337
Barrangou R, Fremaux C, Deveau H (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. https://doi.org/10.1126/science.1138140
Hassan MM, Zhang Y, Yuan G (2021) Construct design for CRISPR/Cas-based genome editing in plants. Trends Plant Sci 26(11):1133–1152. https://doi.org/10.1016/j.tplants.2021.06.015
Doench G, Hartenian E, Graham DB (2014) Rational design of highly active sgRNAs for CRISPR-Cas9- mediated gene inactivation. Nat Biotechnol 32:1262. https://doi.org/10.1038/nbt.3026
Doench JG, Fusi N, Sullender NM (2016) Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPRCas9. Nat Biotechnol 34:184. https://doi.org/10.1038/nbt.3437
Xu H, Xiao T, Chen C (2015) Sequence determinants of improved CRISPR sgRNA design. Genome Res 25:1147–1157. https://doi.org/10.1101/gr.191452.115
Moreno-Mateos MA, Vejnar CE, Beaudoin JD (2015) CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods 12:982. https://doi.org/10.1038/nmeth.3543
Stemmer M, Thumberger T, Del Sol KM (2015) CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE 24:e0124633. https://doi.org/10.1371/journal.pone.0124633
Bae S, Kweon J, Kim HS, Kim JS (2014) Microhomology-based choice of Cas9 nuclease target sites. Nat Methods 11:705–706. https://doi.org/10.1038/nmeth.3015
Kuan PF, Powers S, He S (2017) A systematic evaluation of nucleotide properties for CRISPR sgRNA design. BMC Bioinform 18:297. https://doi.org/10.1186/s12859-017-1697-6
Malina A, Cameron CJ, Robert F (2015) PAM multiplicity marks genomic target sites as inhibitory to CRISPR-Cas9 editing. Nat Commun 6:10124. https://doi.org/10.1038/ncomms10124
Graf R, Li X, Chu VT, Rajewsky K (2019) sgRNA sequence motifs blocking efficient CRISPR/Cas9-mediated gene editing. Cell Rep 26:1098-1103.e3. https://doi.org/10.1016/j.celrep.2019.01.024
Wong N, Liu W, Wang X (2015) WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/ Cas9 system. Genome Biol 16:218. https://doi.org/10.1186/s13059-015-0784-0
Liang G, Zhang H, Lou D, Yu D (2016) Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci Rep 6:1–8. https://doi.org/10.1038/srep21451
Chari R, Yeo NC, Chavez A, Church GM (2017) sgRNA scorer 2.0: a species-independent model to predict CRISPR/Cas9 activity. ACS Synth Biol 6(5):902–904. https://doi.org/10.1021/acssynbio.6b00343
Lin Y, Cradick TJ, Brown MT (2014) CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res 42:7473–7485. https://doi.org/10.1093/nar/gku402
Mendoza BJ, Trinh CT (2018) Enhanced guide-RNA design and targeting analysis for precise CRISPR genome editing of single and consortia of industrially relevant and non-model organisms. Bioinformatics 34:16–23. https://doi.org/10.1093/bioinformatics/btx564
Yue J, Hong C, Wei P (2020) How to start your monocot CRISPR/Cas project: plasmid design, efficiency detection, and offspring analysis. Rice 13:1–13. https://doi.org/10.1186/s12284-019-0354-2
Miki D, Zhang W, Zeng W (2018) CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat Commun 9:1967. https://doi.org/10.1038/s41467-018-04416-0
Zhang Z, Mao Y, Ha S (2016) A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep 35:1519–1533. https://doi.org/10.1007/s00299-015-1900-z
Kurata M, Wolf NK, Lahr WS (2018) Highly multiplexed genome engineering using CRISPR/Cas9 gRNA arrays. PLoS ONE 13:e0198714. https://doi.org/10.1371/journal.pone.0198714
Zhang H, Zhang J, Wei P (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12:797–807. https://doi.org/10.1111/pbi.12200
Zhan X, Lu Y, Zhu JK, Botella JR (2021) Genome editing for plant research and crop improvement. J Integr Plant Biol 63:3–33. https://doi.org/10.1111/jipb.13063
Feng Z, Mao Y, Xu N (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 111:4632–4637. https://doi.org/10.1073/pnas.1400822111
Wang Z, Xing H, Li D (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16:1–12. https://doi.org/10.1186/s13059-015-0715-0
Mao Y, Zhang Z, Feng Z (2016) Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J 14:519–532. https://doi.org/10.1111/pbi.12468
Mao Y, Yang X, Zhou Y (2018) Manipulating plant RNA-silencing pathways to improve the gene editing efficiency of CRISPR/Cas9 systems. Genome Biol 19:1–15. https://doi.org/10.1186/s13059-018-1529-7
Endo A, Masafumi M, Kaya H, Toki S (2016) Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci Rep 6:38169. https://doi.org/10.1038/srep38169
Cermak T, Curtin SJ, Gil-Humanes J (2017) A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29:1196–1217. https://doi.org/10.1105/tpc.16.00922
Tang X, Sretenovic S, Ren Q (2020) Plant prime editors enable precise gene editing in rice cells. Mol Plant 13:667–670. https://doi.org/10.1016/j.molp.2020.03.010
Hua K, Jiang Y, Tao X, Zhu JK (2020) Precision genome engineering in rice using prime editing system. Plant Biotechnol J 18:2167–2169. https://doi.org/10.1111/pbi.13395
Cermak T, Baltes NJ, Cegan R (2015) High-frequency, precise modification of the tomato genome. Genome Biol 16:232. https://doi.org/10.1186/s13059-015-0796-9
Aman R, Ali Z, Butt H (2018) RNA virus interference via CRISPR/ Cas13a system in plants. Genome Biol 19:1–9. https://doi.org/10.1186/s13059-017-1381-1
Lu Y, Tian Y, Shen R (2020) Targeted, efficient sequence insertion and replacement in rice. Nat Biotechnol 38:1402–1407. https://doi.org/10.1038/s41587-020-0581-5
Endo M, Mikami M, Toki S (2015) Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol 56:41–47. https://doi.org/10.1093/pcp/pcu154
Stevens R, Grelon M, Vezon D (2004) A CDC45 homolog in Arabidopsis is essential for meiosis, as shown by RNA interference einduced gene silencing. Plant Cell 16:99–113. https://doi.org/10.1105/tpc.016865
Yan L, Wei S, Wu Y (2015) High efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant 8:1820–1823. https://doi.org/10.1016/j.molp.2015.10.004
Ordon J, Bressan M, Kretschmer C, Dall’Osto L, Marillonnet S, Bassi R, Stuttmann J (2020) Optimized Cas9 expression systems for highly efficient Arabidopsis genome editing facilitate isolation of complex alleles in a single generation. Funct Integr 20:151–162. https://doi.org/10.1007/s10142-019-00665-4
Peng FN, Zhang WX, Zeng WJ, Zhu JK, Miki D (2020) Gene targeting in Arabidopsis via an all-in-one strategy that uses a translational enhancer to aid Cas9 expression. Plant Biotechnol J 18:892–894. https://doi.org/10.1111/pbi.13265
Johnson LM, Du J, Hale CJ (2014) SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507:124–128. https://doi.org/10.1038/nature12931
Papikian A, Liu W, Gallego-Bartolome J, Jacobsen SE (2019) Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat Commun 10:729. https://doi.org/10.1038/s41467-019-08736-7
Gallego-Bartolome J, Gardiner J, Liu W (2018) Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc Natl Acad Sci USA 115:E2125–E2134. https://doi.org/10.1073/pnas.1716945115
Ji L, Jordan WT, Shi X (2018) TET-mediated epimutagenesis of the Arabidopsis thaliana methylome. Nat Commun 9:895. https://doi.org/10.1038/s41467-018-03289-7
Wang X, Ye L, Lyu M (2020) An inducible genome editing system for plants. Nat Plants 6:766–772. https://doi.org/10.1038/s41477-020-0695-2
Decaestecker W, Buono RA, Pfeiffer ML (2019) CRISPR-TSKO: a technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell 31(12):2868–2887. https://doi.org/10.1105/tpc.19.00454
Vazquez-Vilar M, Garcia-Carpintero V, Selma (2021) The GB4.0 platform, an all-in-one tool for CRISPR/Cas-based multiplex genome engineering in plants. Front Plant Sci 12:689937. https://doi.org/10.3389/fpls.2021.689937
Zhang D, Zhang Z, Zhang UT (2021) CRISPR/Cas: A powerful tool for gene function study and crop improvement. J Adv Res 29:207–221. https://doi.org/10.1016/j.jare.2020.10.003
Zhou H, Liu B, Weeks DP, Spalding MH, Yang B (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nuc Acids Res 42:10903–10914. https://doi.org/10.1093/nar/gku806
Saika H, Mori A, Endo M, Toki S (2019) Targeted deletion of rice retrotransposon Tos17 via CRISPR/Cas9. Plant Cell Rep 38:455–458. https://doi.org/10.1007/s00299-018-2357-7
Gao J, Wang G, Ma S (2015) CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol 87:99–110. https://doi.org/10.1007/s11103-014-0263-0
Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112:3570–3575. https://doi.org/10.1073/pnas.1420294112
Lowder LG, Zhang D, Baltes NJ (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169:971–985. https://doi.org/10.1104/pp.15.00636
Cody WB, Scholthof HB, Mirkov TE (2017) Multiplexed gene editing and protein overexpression using a tobacco mosaic virus viral vector. Plant Physiol 17:23–35. https://doi.org/10.1104/pp.17.00411
Miladinovic D, Antunes D, Yildirim K (2021) Targeted plant improvement through genome editing: from laboratory to field. Plant Cell Rep 40:935–951. https://doi.org/10.1007/s00299-020-02655-4
Doll NM, Gilles LM, Gérentes MF (2019) Single and multiple gene knockouts by CRISPR-Cas9 in maize. Plant Cell Rep 38(2019):487–501. https://doi.org/10.1007/s00299-019-02378-1
Bollier N, Buono RA, Jacobs TB, Nowack MK (2020) Efficient simultaneous mutagenesis of multiple genes in specific plant tissues by multiplex CRISPR. Plant Biotech J 19:651–653. https://doi.org/10.1101/2020.11.13.381046
Li R, Li R, Li X (2018) Multiplexed CRISPR/Cas9-mediated metabolic engineering of γ-aminobutyric acid levels in Solanum lycopersicum. Plant Biotech J 16:415–427. https://doi.org/10.1111/pbi.12781
Li X, Wang Y, Chen S (2018) Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front Plant Sci 9:559. https://doi.org/10.3389/fpls.2018.00559
Li Z, Cheng Q, Gan Z (2020) Multiplex CRISPR/Cas9-mediated knockout of soybean LNK2 advances flowering time. Crop J 9:767–776. https://doi.org/10.1016/j.cj.2020.09.005
Reem NT, Van Eck J (2019) Application of CRISPR/Cas9-mediated gene editing in tomato. In: Qi Y (ed) Plant genome editing with CRISPR systems methods and protocols. Springer, Switzerland, pp 171–182
Toda E, Koiso N, Takebayashi A (2019) An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice. Nat Plants 5:363–368. https://doi.org/10.1038/s41477-019-0386-z
Upadhyay SK, Kumar J, Alok A, Tuli R (2013) RNA-guided genome editing for target gene mutations in wheat. Genes Genom Genet 3:2233–2238. https://doi.org/10.1534/g3.113.008847
Xing S, Jia M, Wei L (2018) CRISPR/Cas9-introduced single and multiple mutagenesis in strawberry. J Genet Genomics 45:685–687. https://doi.org/10.1016/j.jgg.2018.04.006
Raitskin O, Patron NJ (2016) Multi-gene engineering in plants with RNA-guided Cas9 nuclease. Curr Opin Biotechnol 37:69–75. https://doi.org/10.1016/j.copbio.2015.11.008
Fan Y, Xin S, Dai X (2020) Efficient genome editing of rubber tree (Hevea brasiliensis) protoplasts using CRISPR/Cas9 ribonucleoproteins. Ind Crops Prod 146:112146. https://doi.org/10.1016/j.indcrop.2020.112146
Woo JW, Kim J, Kwon SI (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33:1162–1164. https://doi.org/10.1038/nbt.3389
Andersson M, Turesson H, Olsson N (2018) Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol Plant 164:378–384. https://doi.org/10.1111/ppl.12731
Chang KS, Kim J, Park H (2020) Enhanced lipid productivity in AGP knockout marine microalga Tetraselmis sp. using a DNA-free CRISPR-Cas9 RNP method. Bioresour Technol 303:122932. https://doi.org/10.1016/j.biortech.2020.122932
Sant’Ana R, Caprestano CA, Nodari RO, Agapito-Tenfen SZ (2020) PEG-delivered CRISPR-Cas9 ribonucleoproteins system for gene-editing screening of maize protoplasts. Genes 11:1029. https://doi.org/10.3390/genes11091029
Ghogare R, Ludwig Y, Bueno GM (2021) Genome editing reagent delivery in plants. Transgenic Res 30:321–335. https://doi.org/10.1007/s11248-021-00239-w
Zhang S, Shen J, Li D, Cheng Y (2021) Strategies in the delivery of Cas9 ribonucleoprotein for CRISPR/Cas9 genome editing. Theranostics 11:614–648. https://doi.org/10.7150/thno.47007
Banakar R, Eggenberger AL, Lee K (2019) High-frequency random DNA insertions upon co-delivery of CRISPR-Cas9 ribonucleoprotein and selectable marker plasmid in rice. Sci Rep 9:19902. https://doi.org/10.1038/s41598-019-55681-y
Liang Z, Chen K, Li T (2017) Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8:14261. https://doi.org/10.1038/ncomms14261
Kim K, Choi J, Won KH (2020) A stable DNA-free screening system for CRISPR/RNPs-mediated gene editing in hot and sweet cultivars of Capsicum annuum. BMC Plant Biol 20:449. https://doi.org/10.1186/s12870-020-02665-0
Liu W, Rudis MR, Cheplick MH (2020) Lipofection-mediated genome editing using DNA-free delivery of the Cas9/gRNA ribonucleoprotein into plant cells. Plant Cell Rep 39:245–257. https://doi.org/10.1007/s00299-019-02488-w
Yu J, Tu L, Subburaj S (2020) Simultaneous targeting of duplicated genes in Petunia protoplasts for flower color modification via CRISPR-Cas9 ribonucleoproteins. Plant Cell Rep 40:1037–1045. https://doi.org/10.1007/s00299-020-02593-1
Zhao X, Jayarathna S, Turesson H (2021) Amylose starch with no detectable branching developed through DNA-free CRISPR-Cas9 mediated mutagenesis of two starch branching enzymes in potato. Sci Rep 11:4311. https://doi.org/10.1038/s41598-021-83462-z
Acknowledgements
This manuscript is written by the leadership of the “Plant Science Initiative of Turkey”, a non-governmental organization aiming to bring the scientists together who work in the field of Plant Sciences in Turkey.
Funding
This review received no external funding.
Author information
Authors and Affiliations
Contributions
Conceptualization, EA, KY, MK, CK, UD; investigation, BAY, IÇ, İS; writing-original draft, EA, KY, MK, CK, BAY, IÇ, İS; writing-review and editing, EA, KY, MK, CK, UD; visualization, MK, BAY; supervision, EA, KY. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no confict of interest regarding the publication of this article.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Aksoy, E., Yildirim, K., Kavas, M. et al. General guidelines for CRISPR/Cas-based genome editing in plants. Mol Biol Rep 49, 12151–12164 (2022). https://doi.org/10.1007/s11033-022-07773-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07773-8